An R-package which contains functions to set up risk-adjusted quality control charts in health care.
You can install the released version of vlad from CRAN with:
And the development version from GitHub with:
Load libraries:
Subset the dataset cardiacsurgery into Phase I (first two years) and Phase II (five years) and estimate a risk model based on phaseI.
data("cardiacsurgery", package = "spcadjust")
cardiacsurgery <- cardiacsurgery %>% rename(s = Parsonnet) %>%
mutate(y = ifelse(status == 1 & time <= 30, 1, 0),
phase = factor(ifelse(date < 2*365, "I", "II")))
head(cardiacsurgery)
#> date time status s surgeon y phase
#> 1 1 90 0 15 7 0 I
#> 2 1 90 0 9 3 0 I
#> 3 2 90 0 2 5 0 I
#> 4 3 90 0 8 7 0 I
#> 5 3 90 0 7 1 0 I
#> 6 3 90 0 40 1 0 I
phaseI <- filter(cardiacsurgery, phase == "I") %>% select(s, y)
coeff <- round(coef(glm(y ~ s, data = phaseI, family = "binomial")), 3)
print(coeff)
#> (Intercept) s
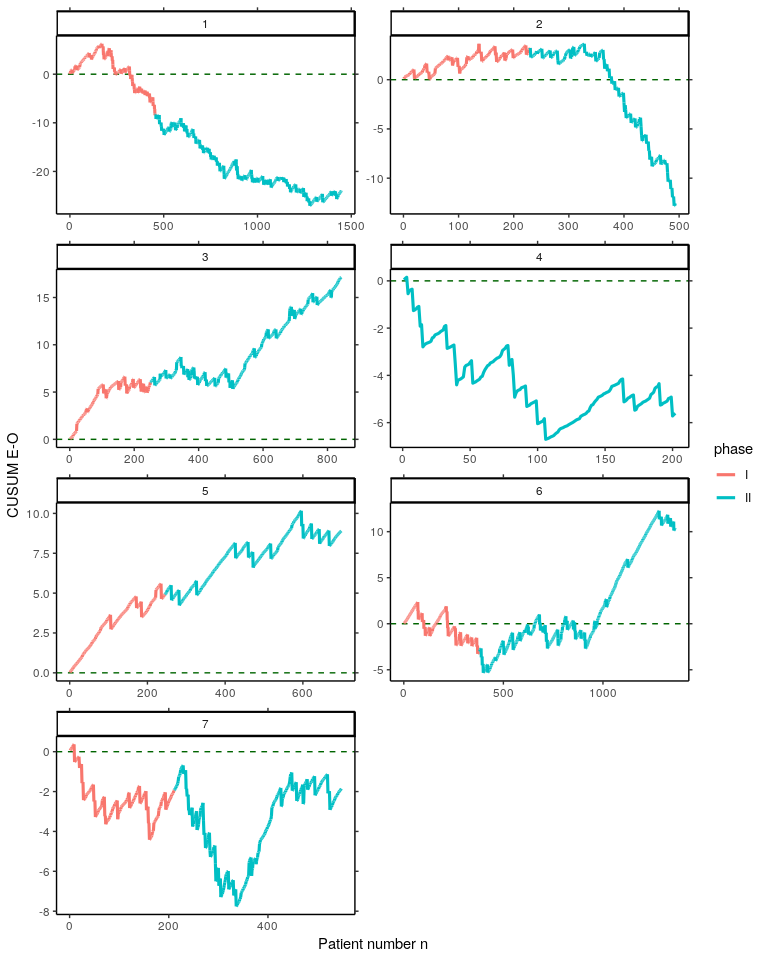
#> -3.79 0.08By using the estimated risk model coefficients coeff, for each pair of Parsonnet score s and operation outcome values y, the difference between expected and observed outcome is calculated with the function calceo(). Thereafter, differences are cummulated to create the VLAD. This is done for all seven surgeons of the cardiacsurgery dataset. Results are saved to the object vlads7.
vlads7 <- lapply(1:7, function(j){
Si <- filter(cardiacsurgery, surgeon == j)
EO <- sapply(seq_along(Si$s), function(i) calceo(df = Si[i, c("s", "y")], coeff = coeff))
select(Si, surgeon, phase) %>% mutate(n = 1:length(EO), cEO = cumsum(EO))
}) Create Variable life-adjusted Displays for each surgeon from the object vlads7.
vlads7 %>%
bind_rows() %>%
gather(key = "Surgeon", value = value, c(-n, -surgeon, -phase)) %>%
ggplot(aes(x = n, y = value, colour = phase, group = Surgeon)) +
geom_hline(yintercept = 0, colour = "darkgreen", linetype = "dashed") +
geom_line(size = 1.1) + facet_wrap( ~ surgeon, ncol = 2, scales = "free") +
labs(x="Patient number n", y="CUSUM E-O") + theme_classic() +
scale_y_continuous(sec.axis = dup_axis(name = NULL, labels = NULL)) +
scale_x_continuous(sec.axis = dup_axis(name = NULL, labels = NULL))
S2 <- filter(cardiacsurgery, surgeon == 2) %>% select(phase, s, y)
S2I <- subset(S2, c(phase == "I"))
S2II <- subset(S2, c(phase == "II"))
coeff <- coef(glm(y ~ s, data = S2I, family = "binomial"))
EO <- sapply(1:nrow(S2), function(i) calceo(df = S2[i, c("s", "y")], coeff = coeff))
df1 <- select(S2, phase) %>% mutate(n = row_number(), cEO = cumsum(EO))
df2 <- gather(df1, variable, value, c(-n, -phase))
p1 <- ggplot(df2, aes(x = n, y = value, colour = phase)) +
geom_hline(yintercept = 0, linetype = "dashed") + geom_line() + geom_point() +
labs(x = "Patient number", y = "CUSUM E-O") + theme_classic() +
scale_y_continuous(sec.axis = dup_axis(name = NULL, labels = NULL)) +
scale_x_continuous(sec.axis = dup_axis(name = NULL, labels = NULL))
p1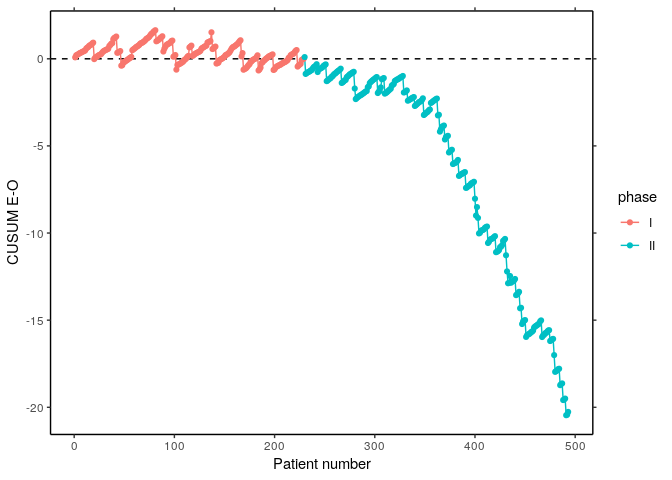
Upper and lower control limits of the risk-adjusted CUSUM chart based on log-likelihood ratio statistic can be computed with the function racusum_arl_h_sim(). The implemention uses parallel simulation and a multi-stage search procedure.
# set a random number generator for parallel computations
RNGkind("L'Ecuyer-CMRG")
# number of simulation runs
m <- 10^4
# assign cores
nc <- parallel::detectCores()
# verbose calculation
UCL_sim <- racusum_crit_sim(L0 = 740, df = S2I[, c("s", "y")], coeff = coeff, m = m, RA = 2, nc = nc, verbose = TRUE)
#> h = 1 ARL = 75.006
#> h = 2 ARL = 383.3554
#> h = 3 ARL = 1312.8564
#> h = 2.9 ARL = 1181.3768
#> h = 2.8 ARL = 1052.3355
#> h = 2.7 ARL = 928.6587
#> h = 2.6 ARL = 822.3063
#> h = 2.5 ARL = 730.0184
#> h = 2.51 ARL = 739.8392
#> h = 2.52 ARL = 747.3412
#> h = 2.519 ARL = 746.4942
#> h = 2.518 ARL = 745.9933
#> h = 2.517 ARL = 745.6282
#> h = 2.516 ARL = 744.6935
#> h = 2.515 ARL = 744.0614
#> h = 2.514 ARL = 743.2221
#> h = 2.513 ARL = 742.2656
#> h = 2.512 ARL = 741.7172
#> h = 2.511 ARL = 741.3188
#> h = 2.51 ARL = 739.8392
#> h = 2.5101 ARL = 739.862
#> h = 2.5102 ARL = 739.862
#> h = 2.5103 ARL = 740.0315
# quite calculation
LCL_sim <- racusum_crit_sim(L0 = 740, df = S2I[, c("s", "y")], coeff = coeff, m = m, RA = 1/2, nc = nc, verbose = FALSE)
round(cbind(UCL_sim, LCL_sim), 3)
#> UCL_sim LCL_sim
#> [1,] 2.51 2.281Wittenberg et al. (2018). A simple signaling rule for variable life-adjusted display derived from an equivalent risk-adjusted CUSUM chart
Steiner et al. (2000). Monitoring surgical performance using risk-adjusted cumulative sum charts
Philipp Wittenberg and Sven Knoth
GPL (>= 2)