# Install devtools if not previously installed.
# install.packages("devtools")
# If there are errors (converted from warning) during installation related to packages built under different version of R,
# they can be ignored by setting the environment variable R_REMOTES_NO_ERRORS_FROM_WARNINGS="true" before calling install_github()
Sys.setenv(R_REMOTES_NO_ERRORS_FROM_WARNINGS="true")
devtools::install_github("jameswcraig/tidyvpc")library(magrittr)
library(ggplot2)
library(tidyvpc)
# Filter MDV = 0
obs_data <- as.data.table(tidyvpc::obs_data)[MDV == 0]
sim_data <- as.data.table(tidyvpc::sim_data)[MDV == 0]
#Add LLOQ for each Study
obs_data$LLOQ <- obs_data[, ifelse(STUDY == "Study A", 50, 25)]
# Binning Method on x-variable (NTIME)
vpc <- observed(obs_data, x=TIME, y=DV) %>%
simulated(sim_data, y=DV) %>%
censoring(blq=(DV < LLOQ), lloq=LLOQ) %>%
stratify(~ STUDY) %>%
binning(bin = NTIME) %>%
vpcstats()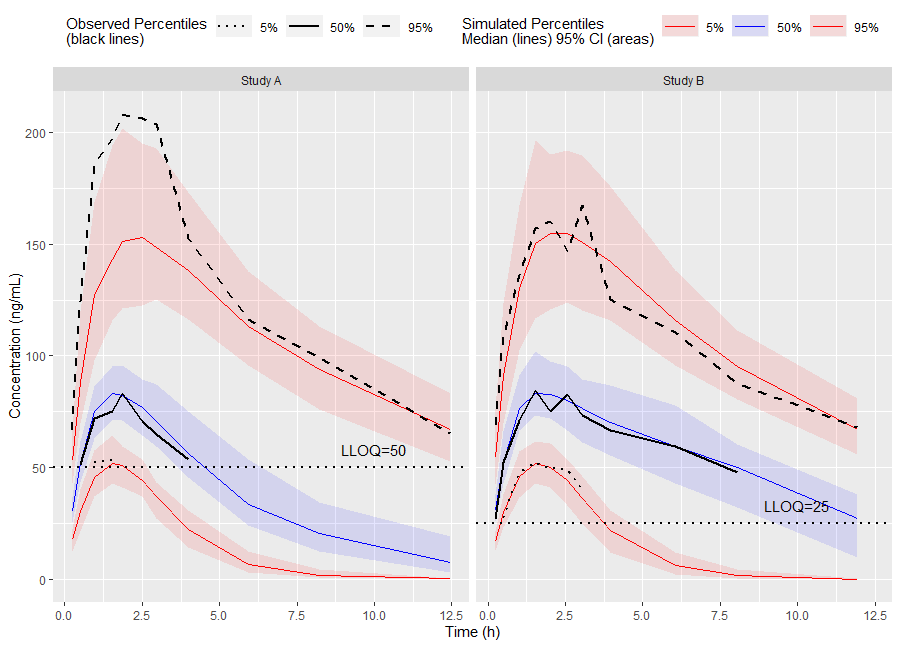
Plot Code:
ggplot(vpc$stats, aes(x=xbin)) +
facet_grid(~ STUDY) +
geom_ribbon(aes(ymin=lo, ymax=hi, fill=qname, col=qname, group=qname), alpha=0.1, col=NA) +
geom_line(aes(y=md, col=qname, group=qname)) +
geom_line(aes(y=y, linetype=qname), size=1) +
geom_hline(data=unique(obs_data[, .(STUDY, LLOQ)]),
aes(yintercept=LLOQ), linetype="dotted", size=1) +
geom_text(data=unique(obs_data[, .(STUDY, LLOQ)]),
aes(x=10, y=LLOQ, label=paste("LLOQ", LLOQ, sep="="),), vjust=-1) +
scale_colour_manual(
name="Simulated Percentiles\nMedian (lines) 95% CI (areas)",
breaks=c("q0.05", "q0.5", "q0.95"),
values=c("red", "blue", "red"),
labels=c("5%", "50%", "95%")) +
scale_fill_manual(
name="Simulated Percentiles\nMedian (lines) 95% CI (areas)",
breaks=c("q0.05", "q0.5", "q0.95"),
values=c("red", "blue", "red"),
labels=c("5%", "50%", "95%")) +
scale_linetype_manual(
name="Observed Percentiles\n(black lines)",
breaks=c("q0.05", "q0.5", "q0.95"),
values=c("dotted", "solid", "dashed"),
labels=c("5%", "50%", "95%")) +
guides(
fill=guide_legend(order=2),
colour=guide_legend(order=2),
linetype=guide_legend(order=1)) +
theme(
legend.position="top",
legend.key.width=grid::unit(1, "cm")) +
labs(x="Time (h)", y="Concentration (ng/mL)")Or use the built in plot() function from the tidyvpc package.
# Binless method using 10%, 50%, 90% quantiles and LOESS Prediction Corrected
# Add PRED variable to observed data from first replicate of sim_data
obs_data$PRED <- sim_data[REP == 1, PRED]
vpc <- observed(obs_data, x=TIME, y=DV) %>%
simulated(sim_data, y=DV) %>%
stratify(~ GENDER) %>%
predcorrect(pred=PRED) %>%
binless(qpred = c(0.1, 0.5, 0.9), loess.ypc = TRUE) %>%
vpcstats()
plot(vpc)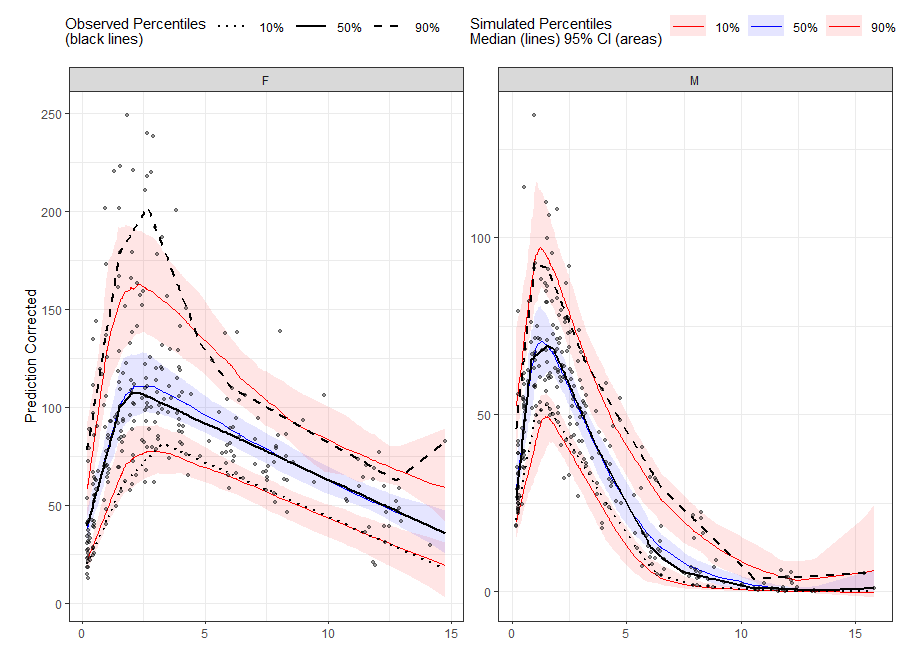
The tidyvpc package contains a wrapper function to install necessary dependencies and run the Shiny-VPC Application. Use the runShinyVPC() function from tidyvpc to parameterize VPC from a GUI and generate correpsponding tidyvpc and ggplot2 code to reproduce VPC in your local R session.
runShinyVPC()Note: Internet access is required to use runShinyVPC()