

Several different sigmoid functions are implemented, including a wrapper function, SoftMax preprocessing and inverse functions.
The stable version can be installed from CRAN using:
The development version, to be used at your peril, can be installed from GitHub using the devtools package.
Following installation, the package can be loaded using:
The sigmoid() function returns the sigmoid value of the input(s), by default this is done using the standard logistic function.
Inputs can also be tensors, such as vectors, matrices, or arrays.
sigmoid(-5:5)
#> [1] 0.006692851 0.017986210 0.047425873 0.119202922 0.268941421
#> [6] 0.500000000 0.731058579 0.880797078 0.952574127 0.982013790
#> [11] 0.993307149
sigmoid( matrix(-3:5,nrow=3) ) # etc.
#> [,1] [,2] [,3]
#> [1,] 0.04742587 0.5000000 0.9525741
#> [2,] 0.11920292 0.7310586 0.9820138
#> [3,] 0.26894142 0.8807971 0.9933071The sigmoid() function is a wrapper, which by default uses the logistic() function, it can also use other methods.
sigmoid( -5:5, method='Gompertz' )
#> [1] 3.507389e-65 1.942338e-24 1.892179e-09 6.179790e-04 6.598804e-02
#> [6] 3.678794e-01 6.922006e-01 8.734230e-01 9.514320e-01 9.818511e-01
#> [11] 9.932847e-01These functions can also be accessed directly.
logistic(1:5)
#> [1] 0.7310586 0.8807971 0.9525741 0.9820138 0.9933071
Gompertz(-1:-5)
#> [1] 6.598804e-02 6.179790e-04 1.892179e-09 1.942338e-24 3.507389e-65These mappings are similar but not identical.
library(ggplot2)
input = -5:5
df = data.frame(input, logistic(input), Gompertz(input))
ggplot(df, aes(input, logistic(input))) + geom_line() +
geom_line(aes(input,Gompertz(input)), colour='red')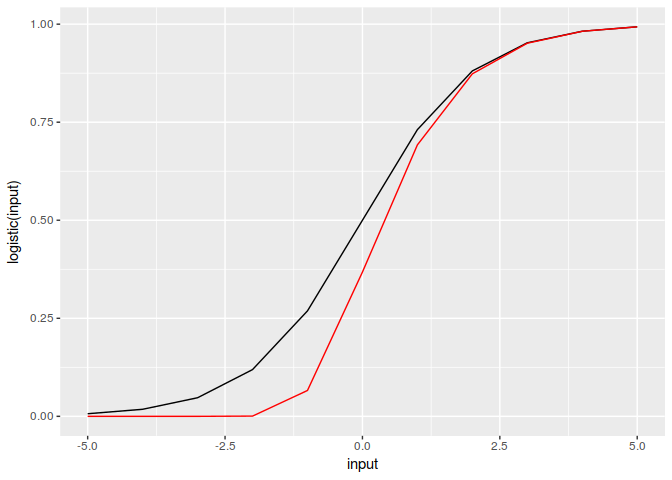
For inverses, additional parameters, SoftMax, etc. see the vignette.
vignette('sigmoid')For general information on using the package, please refer to the help files.
An overview of the changes is available in the NEWS file.
There is a dedicated website with information hosted on my personal website.
Development takes place on the GitHub page.
http://github.com/bquast/sigmoid
Bugs can be filed on the issues page on GitHub.