

Genomic alterations including single nucleotide substitution (SBS), copy number alteration (CNA), etc. are the major force for cancer initialization and development. Due to the specificity of molecular lesions caused by genomic alterations, we can generate characteristic alteration spectra, called ‘signature’. This package helps users to extract, analyze and visualize signatures from genomic alteration records, thus providing new insight into cancer study.
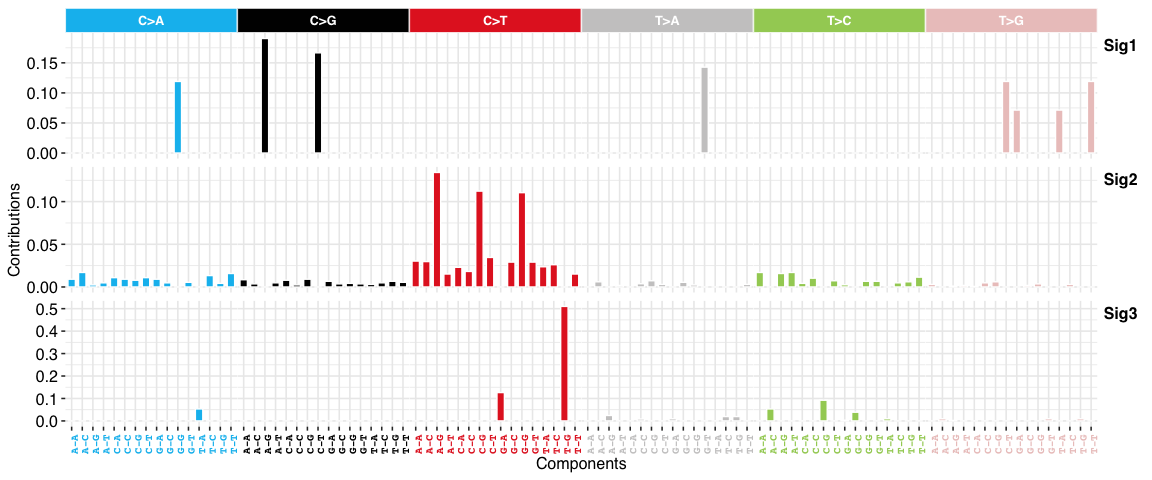
SBS signatures:
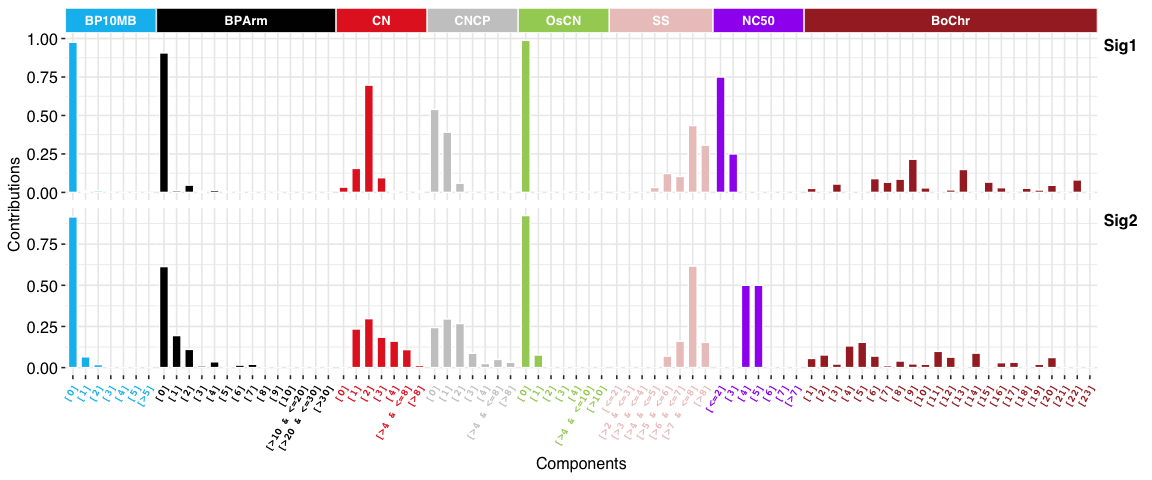
Copy number signatures:
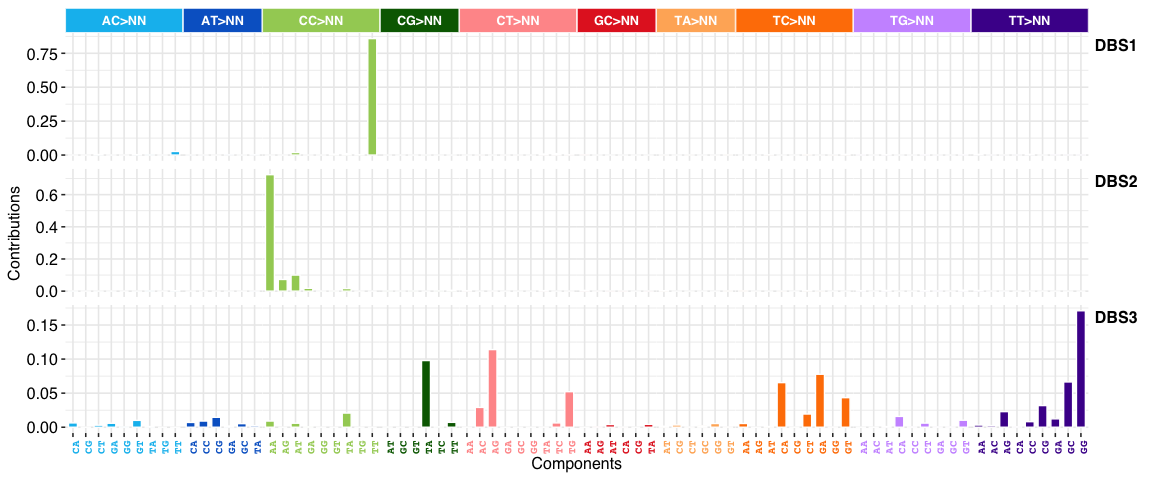
DBS signatures:
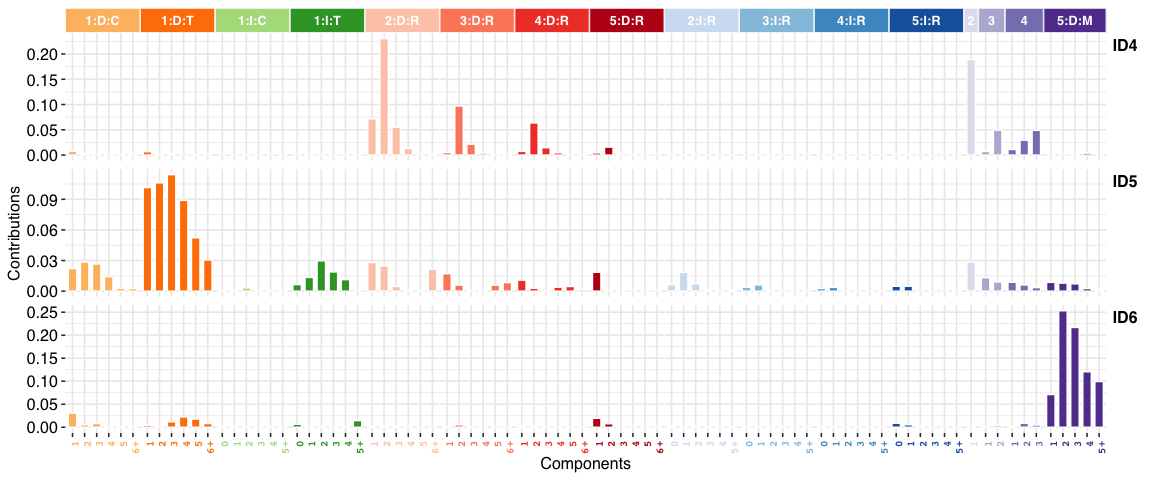
INDEL (i.e. ID) signatures:
You can install the stable release of sigminer from CRAN with:
install.packages("sigminer", dependencies = TRUE)
# Or
BiocManager::install("sigminer", dependencies = TRUE)You can install the development version of sigminer from Github with:
remotes::install_github("ShixiangWang/sigminer", dependencies = TRUE)
# For Chinese users, run
remotes::install_git("https://gitee.com/ShixiangWang/sigminer", dependencies = TRUE)A complete documentation of sigminer can be read online at https://shixiangwang.github.io/sigminer-doc/ (For Chinese users, you can also read it at https://shixiangwang.gitee.io/sigminer-doc). All functions are well organized and documented at https://shixiangwang.github.io/sigminer/reference/index.html (For Chinese users, you can also read it at https://shixiangwang.gitee.io/sigminer/reference/index.html). For usage of a specific function fun, run ?fun in your R console to see its documentation.
Wang, Shixiang, et al. “Copy number signature analyses in prostate cancer reveal distinct etiologies and clinical outcomes” medRxiv (2020) https://www.medrxiv.org/content/early/2020/04/29/2020.04.27.20082404
If you use NMF package in R, please also cite:
Gaujoux, Renaud, and Cathal Seoighe. "A Flexible R Package for
Nonnegative Matrix Factorization."" BMC Bioinformatics 11, no. 1 (December 2010).The method “M” for extracting copy number signatures was based in part on the source code from paper Copy number signatures and mutational processes in ovarian carcinoma, if you use this feature, please also cite:
Macintyre, Geoff, et al. "Copy number signatures and mutational
processes in ovarian carcinoma." Nature genetics 50.9 (2018): 1262.The code for extracting SBS signatures was based in part on the source code of the maftools package, if you use this feature, please also cite:
Mayakonda, Anand, et al. "Maftools: efficient and comprehensive analysis
of somatic variants in cancer." Genome research 28.11 (2018): 1747-1756.The code for extracting mutational signatures was based in part on the source code of the SignatureAnalyzer package, if you use this feature, please also cite:
Kim, Jaegil, et al. "Somatic ERCC2 mutations are associated with a distinct genomic
signature in urothelial tumors." Nature genetics 48.6 (2016): 600.The software is made available for non commercial research purposes only under the MIT. However, notwithstanding any provision of the MIT License, the software currently may not be used for commercial purposes without explicit written permission after contacting Shixiang Wang wangshx@shanghaitech.edu.cn or Xue-Song Liu liuxs@shanghaitech.edu.cn.
MIT © 2019-2020 Shixiang Wang, Xue-Song Liu
MIT © 2018 Geoffrey Macintyre
MIT © 2018 Anand Mayakonda
Cancer Biology Group @ShanghaiTech
Research group led by Xue-Song Liu in ShanghaiTech University