

The goal of pedtools is to provide a lightweight, but comprehensive tool set for creating, manipulating and visualizing pedigrees with or without marker data. Common pedigree structures are quickly produced with tailor-made functions, while a range of utilities enable modifications like adding or removing individuals, extracting subsets, loop breaking, and merging pedigrees. The plotting functionality is imported from the kinship2 package.
pedtools is a continuation of the paramlink package, which is no longer actively developed.
To get pedtools, install from CRAN as follows:
Alternatively, you can obtain the latest development version from GitHub:
# install.packages("devtools") # install devtools if needed
devtools::install_github("magnusdv/pedtools")The following example illustrates how pedigrees and markers may be built from scratch.
library(pedtools)
# Create pedigree
x = cousinPed(degree = 0, removal = 2)
x = addChildren(x, father = 3, nch = 2, sex = 2)
# Relabel according to plot order
x = relabel(x, "asPlot")
# Create marker and attach to pedigree
m = marker(x, "7" = "a/b", "11" = "b/b")
# Plot pedigree with genotypes
plot(x, marker = m, hatched = leaves(x))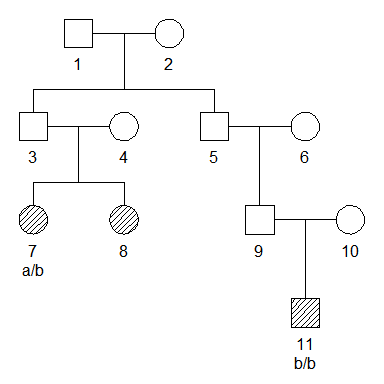
For details about what pedtools can do, and many other examples, the vignette is the recommended place to start.