To get pedmut, install from CRAN as follows:
Alternatively, you can obtain the latest development version from GitHub:
The pedmut package aims to provide a framework for modeling mutations in pedigree computations. Although the package is self-contained, its main purpose is to be imported by other packages, like pedprobr and forrel, in applications involving pedigree likelihoods.
To run the examples below, load pedmut and pedprobr.
Consider the situation shown in the figure below, where father and son are homozygous for different alleles at an autosomal marker with 4 alleles (1,2,3,4). This is a Mendelian error, and gives a likelihood of 0 unless mutations are modelled.
The following code creates the pedigree and the marker, using a “proportional” model for mutations, and computes the likelihood.
# Create pedigree
x = nuclearPed(father = "fa", mother = "mo", child = "boy")
# Create marker with mutation model
m = marker(x, fa = 1, boy = 2, alleles = 1:4, mutmod = "prop", rate = 0.1)
# Plot
plot(x, marker = m)
# Compute likelihood
likelihood(x, m)
#> [1] 0.0005208333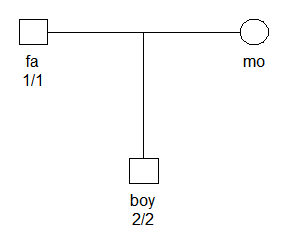
In the above code pedmut is involved twice: first in marker(), translating the arguments mutmod = "prop" and rate = 0.1 into a complete mutation model. And secondly inside likelihood(), by processing the mutation model in setting up the likelihood calculation.
To see details about the mutation model attached to a marker, we can use the mutmod() accessor:
mutmod(m)
#> Unisex mutation matrix:
#> 1 2 3 4
#> 1 0.90000000 0.03333333 0.03333333 0.03333333
#> 2 0.03333333 0.90000000 0.03333333 0.03333333
#> 3 0.03333333 0.03333333 0.90000000 0.03333333
#> 4 0.03333333 0.03333333 0.03333333 0.90000000
#>
#> Model: proportional
#> Rate: 0.1
#> Frequencies: 0.25, 0.25, 0.25, 0.25
#>
#> Stationary: Yes
#> Reversible: Yes
#> Lumpable: AlwaysA mutation matrix is defined in pedmut, as a stochastic matrix with each row summing to 1, where the rows and columns are named with allele labels.
Two central functions of package are mutationMatrix() and mutationModel(). The former of these constructs a single mutation matrix according to various model specifications. The latter is a shortcut for producing what is typically required in practical applications, namely a list of two mutation matrices, named “male” and “female”.
The mutations models currently implemented in pedmut are:
equal: All mutations equally likely; probability 1-rate of no mutation. Parameters: rate.
proportional: Mutation probabilities are proportional to the target allele frequencies. Parameters: rate, afreq.
random: This produces a matrix of random numbers, each row normalised to have sum 1. Parameters: seed.
custom: Allows any valid mutation matrix to be provided by the user. Parameters: matrix.
onestep: Applicable if all alleles are integers. Mutations are allowed only to the nearest integer neighbour. Parameters: rate.
stepwise: For this model alleles must be integers or decimal numbers with a single decimal, such as ‘17.1’, indicating a microvariant. Mutation rates depend on whether transitions are within the same group or not, i.e., between integer alleles and microvariants in the latter case. Mutations also depend on the size of the mutation as modelled by the parameter range, the relative probability of mutating n+1 steps versus mutating n steps. Parameters: rate, rate2, range.
trivial: Diagonal mutation matrix with 1 on the diagonal. Parameters: None.
Certain properties of mutation models are of particular interest - both theoretical and practical - for likelihood computations. The pedmut package provides utility functions for quickly checking whether a given model these properties:
isStationary(M, afreq): Checks if afreq is a right eigenvector of the mutation matrix M
isReversible(M, afreq): Checks if M together with afreq form a reversible Markov chain, i.e., that they satisfy the detailed balance criterion
isLumpable(M, lump): Checks if M allows clustering (“lumping”) of a given subset of alleles. This implements the necessary and sufficient condition of strong lumpability of Kemeny and Snell: Finite Markov Chains, 1960
alwaysLumpable(M): Checks if M allows lumping of any allele subset
An equal model with rate 0.1:
mutationMatrix("equal", rate = 0.1, alleles = 1:3)
#> 1 2 3
#> 1 0.90 0.05 0.05
#> 2 0.05 0.90 0.05
#> 3 0.05 0.05 0.90
#>
#> Model: equal
#> Rate: 0.1
#>
#> Lumpable: AlwaysTo illustrate the stepwise model, we recreate the mutation matrix in Simonsson and Mostad (FSI:Genetics, 2015), Section 2.1.3. This is done as follows:
mutationMatrix(model = "stepwise",
alleles = c("16", "17", "18", "16.1", "17.1"),
rate = 0.003, rate2 = 0.001, range = 0.5)
#> 16 17 18 16.1 17.1
#> 16 0.9960000000 0.0020000000 0.0010000000 0.0005000000 0.0005000000
#> 17 0.0015000000 0.9960000000 0.0015000000 0.0005000000 0.0005000000
#> 18 0.0010000000 0.0020000000 0.9960000000 0.0005000000 0.0005000000
#> 16.1 0.0003333333 0.0003333333 0.0003333333 0.9960000000 0.0030000000
#> 17.1 0.0003333333 0.0003333333 0.0003333333 0.0030000000 0.9960000000
#>
#> Model: stepwise
#> Rate: 0.003
#> Rate2: 0.001
#> Range: 0.5
#>
#> Lumpable: Not alwaysA simpler version of the stepwise model above, is the onestep model, in which only the immediate neighbouring integers are reachable by mutation. This model is only applicable when all alleles are integers.