














ggstatsplot: ggplot2 Based Plots with Statistical Details| Package | Status | Usage | GitHub | References |
|---|---|---|---|---|
 |
 |
|||
 |
 |
|||
 |
 |
 |
 |
|
 |
 |
|||
 |
 |
 |
||
 |
 |

“What is to be sought in designs for the display of information is the clear portrayal of complexity. Not the complication of the simple; rather … the revelation of the complex.”
- Edward R. Tufte
ggstatsplot is an extension of ggplot2 package for creating graphics with details from statistical tests included in the information-rich plots themselves. In a typical exploratory data analysis workflow, data visualization and statistical modeling are two different phases: visualization informs modeling, and modeling in its turn can suggest a different visualization method, and so on and so forth. The central idea of ggstatsplot is simple: combine these two phases into one in the form of graphics with statistical details, which makes data exploration simpler and faster.
It, therefore, produces a limited kinds of plots for the supported analyses:
| Function | Plot | Description |
|---|---|---|
ggbetweenstats |
violin plots | for comparisons between groups/conditions |
ggwithinstats |
violin plots | for comparisons within groups/conditions |
gghistostats |
histograms | for distribution about numeric variable |
ggdotplotstats |
dot plots/charts | for distribution about labeled numeric variable |
ggpiestats |
pie charts | for categorical data |
ggbarstats |
bar charts | for categorical data |
ggscatterstats |
scatterplots | for correlations between two variables |
ggcorrmat |
correlation matrices | for correlations between multiple variables |
ggcoefstats |
dot-and-whisker plots | for regression models and meta-analysis |
In addition to these basic plots, ggstatsplot also provides grouped_ versions (see below) that makes it easy to repeat the same analysis for any grouping variable.
Currently, it supports only the most common types of statistical tests: parametric, nonparametric, robust, and bayesian versions of t-test/anova, correlation analyses, contingency table analysis, meta-analysis, and regression analyses.
The table below summarizes all the different types of analyses currently supported in this package-
| Functions | Description | Parametric | Non-parametric | Robust | Bayes Factor |
|---|---|---|---|---|---|
ggbetweenstats |
Between group/condition comparisons | Yes | Yes | Yes | Yes |
ggwithinstats |
Within group/condition comparisons | Yes | Yes | Yes | Yes |
gghistostats, ggdotplotstats |
Distribution of a numeric variable | Yes | Yes | Yes | Yes |
ggcorrmat |
Correlation matrix | Yes | Yes | Yes | Yes |
ggscatterstats |
Correlation between two variables | Yes | Yes | Yes | Yes |
ggpiestats, ggbarstats |
Association between categorical variables | Yes | NA |
NA |
Yes |
ggpiestats, ggbarstats |
Equal proportions for categorical variable levels | Yes | NA |
NA |
Yes |
ggcoefstats |
Regression model coefficients | Yes | Yes | Yes | Yes |
ggcoefstats |
Random-effects meta-analysis | Yes | No | Yes | Yes |
For all statistical tests reported in the plots, the default template abides by the APA gold standard for statistical reporting. For example, here are results from Yuen’s test for trimmed means (robust t-test):
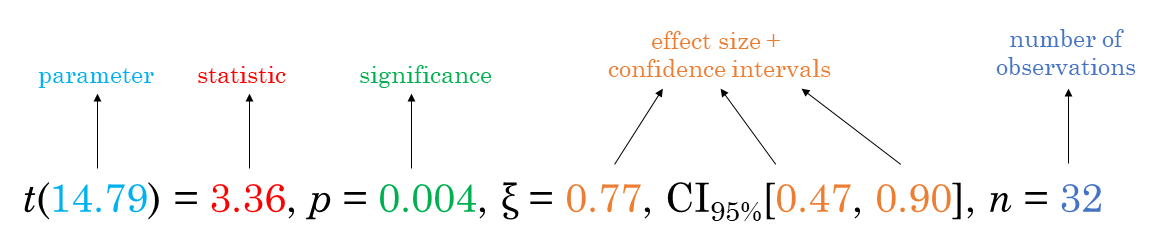
Here is a summary table of all the statistical tests currently supported across various functions: https://indrajeetpatil.github.io/statsExpressions/articles/stats_details.html
To get the latest, stable CRAN release:
Note: If you are on a linux machine, you will need to have OpenGL libraries installed (specifically, libx11, mesa and Mesa OpenGL Utility library - glu) for the dependency package rgl to work.
You can get the development version of the package from GitHub. To see what new changes (and bug fixes) have been made to the package since the last release on CRAN, you can check the detailed log of changes here: https://indrajeetpatil.github.io/ggstatsplot/news/index.html
If you are in hurry and want to reduce the time of installation, prefer-
# needed package to download from GitHub repo
install.packages("remotes")
# downloading the package from GitHub (needs `remotes` package to be installed)
remotes::install_github(
repo = "IndrajeetPatil/ggstatsplot", # package path on GitHub
dependencies = FALSE, # assumes you have already installed needed packages
quick = TRUE # skips docs, demos, and vignettes
)If time is not a constraint-
remotes::install_github(
repo = "IndrajeetPatil/ggstatsplot", # package path on GitHub
dependencies = TRUE, # installs packages which ggstatsplot depends on
upgrade_dependencies = TRUE # updates any out of date dependencies
)If you are not using the RStudio IDE and you get an error related to “pandoc” you will either need to remove the argument build_vignettes = TRUE (to avoid building the vignettes) or install pandoc. If you have the rmarkdown R package installed then you can check if you have pandoc by running the following in R:
If you want to cite this package in a scientific journal or in any other context, run the following code in your R console:
citation("ggstatsplot")
#>
#> Patil, I. (2018). ggstatsplot: 'ggplot2' Based Plots with Statistical
#> Details. CRAN. Retrieved from
#> https://cran.r-project.org/web/packages/ggstatsplot/index.html
#>
#> A BibTeX entry for LaTeX users is
#>
#> @Article{,
#> title = {{ggstatsplot}: 'ggplot2' Based Plots with Statistical Details},
#> author = {Indrajeet Patil},
#> year = {2018},
#> journal = {CRAN},
#> url = {https://CRAN.R-project.org/package=ggstatsplot},
#> doi = {10.5281/zenodo.2074621},
#> }There is currently a publication in preparation corresponding to this package and the citation will be updated once it’s published.
To see the detailed documentation for each function in the stable CRAN version of the package, see:
To see the documentation relevant for the development version of the package, see the dedicated website for ggstatplot, which is updated after every new commit: https://indrajeetpatil.github.io/ggstatsplot/.
Here are examples of the main functions currently supported in ggstatsplot.
Note: If you are reading this on GitHub repository, the documentation below is for the development version of the package. So you may see some features available here that are not currently present in the stable version of this package on CRAN. For documentation relevant for the CRAN version, see: https://CRAN.R-project.org/package=ggstatsplot/readme/README.html
ggbetweenstatsThis function creates either a violin plot, a box plot, or a mix of two for between-group or between-condition comparisons with results from statistical tests in the subtitle. The simplest function call looks like this-
# loading needed libraries
library(ggstatsplot)
# for reproducibility
set.seed(123)
# plot
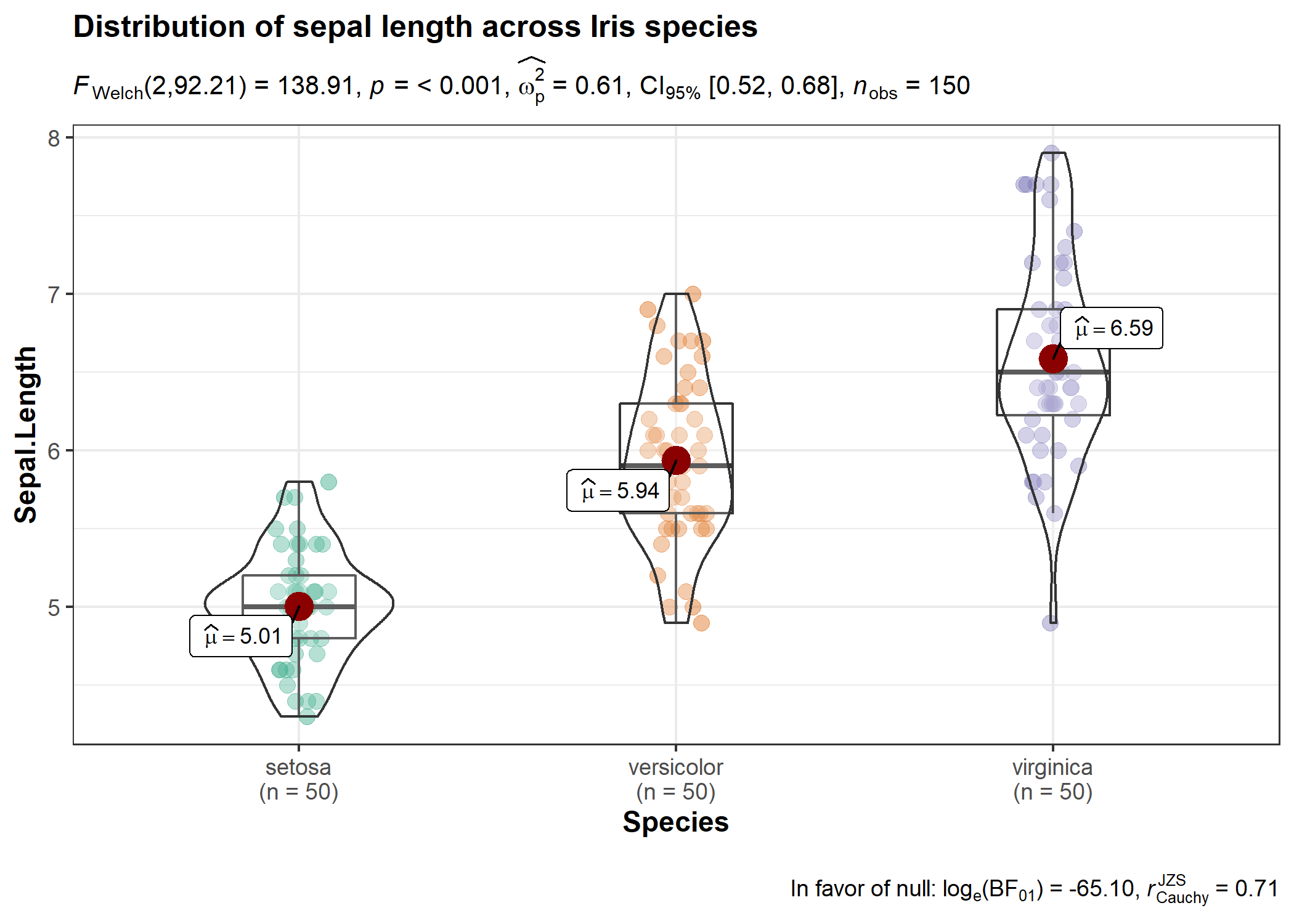
ggstatsplot::ggbetweenstats(
data = iris,
x = Species,
y = Sepal.Length,
title = "Distribution of sepal length across Iris species",
messages = FALSE
)
Note that this function returns object of class ggplot and thus can be further modified using ggplot2 functions.
A number of other arguments can be specified to make this plot even more informative or change some of the default options. Additionally, this time we will use a grouping variable that has only two levels. The function will automatically switch from carrying out an ANOVA analysis to a t-test.
The type (of test) argument also accepts the following abbreviations: "p" (for parametric) or "np" (for nonparametric) or "r" (for robust) or "bf" (for Bayes Factor). Additionally, the type of plot to be displayed can also be modified ("box", "violin", or "boxviolin").
A number of other arguments can be specified to make this plot even more informative or change some of the default options.
# for reproducibility
set.seed(123)
library(ggplot2)
# plot
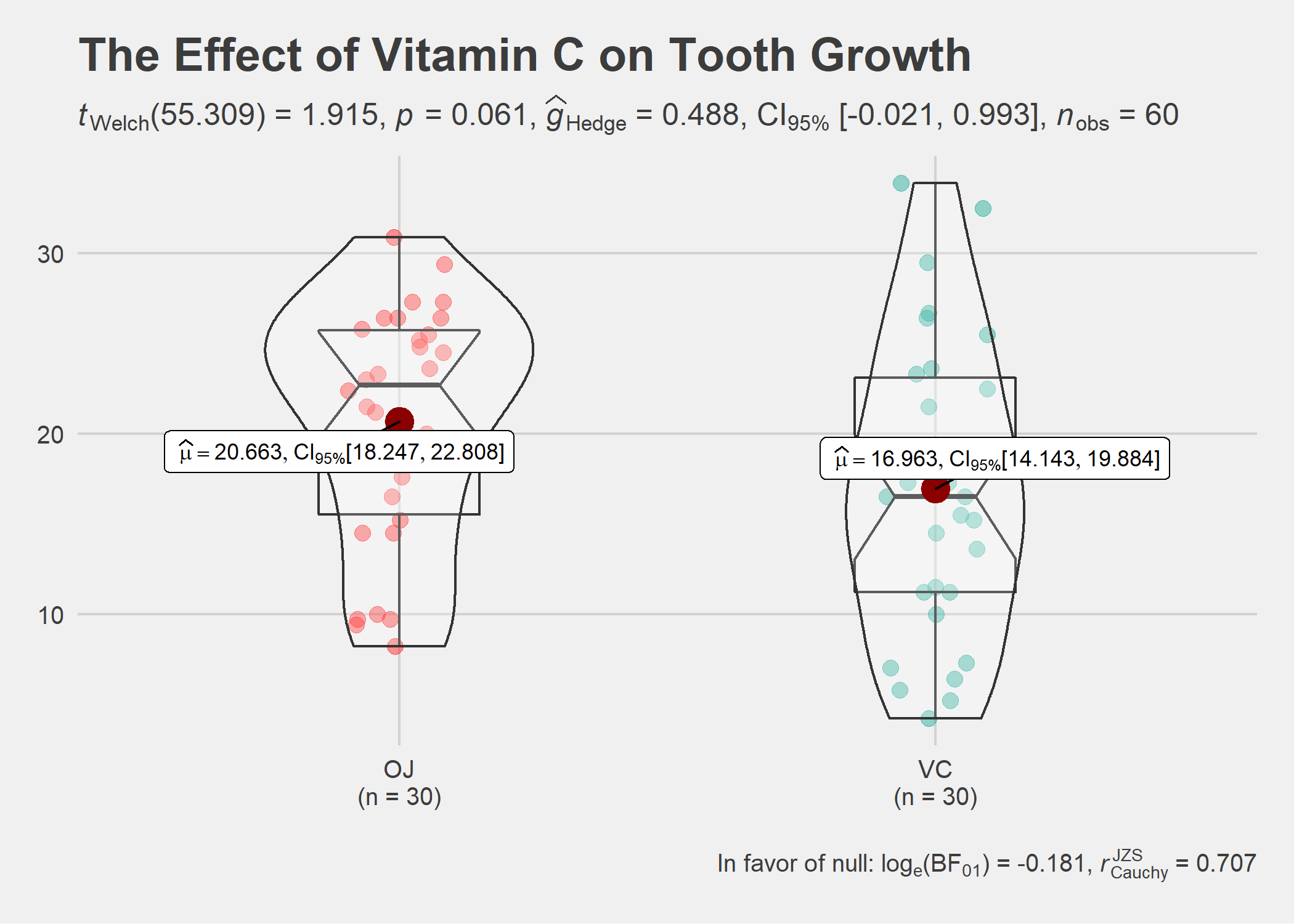
ggstatsplot::ggbetweenstats(
data = ToothGrowth,
x = supp,
y = len,
notch = TRUE, # show notched box plot
mean.ci = TRUE, # whether to display confidence interval for means
k = 3, # number of decimal places for statistical results
outlier.tagging = TRUE, # whether outliers need to be tagged
outlier.label = dose, # variable to be used for the outlier tag
xlab = "Supplement type", # label for the x-axis variable
ylab = "Tooth length", # label for the y-axis variable
title = "The Effect of Vitamin C on Tooth Growth", # title text for the plot
ggtheme = ggthemes::theme_fivethirtyeight(), # choosing a different theme
ggstatsplot.layer = FALSE, # turn off `ggstatsplot` theme layer
package = "wesanderson", # package from which color palette is to be taken
palette = "Darjeeling1", # choosing a different color palette
messages = FALSE
)
Additionally, there is also a grouped_ variant of this function that makes it easy to repeat the same operation across a single grouping variable:
# for reproducibility
set.seed(123)
# plot
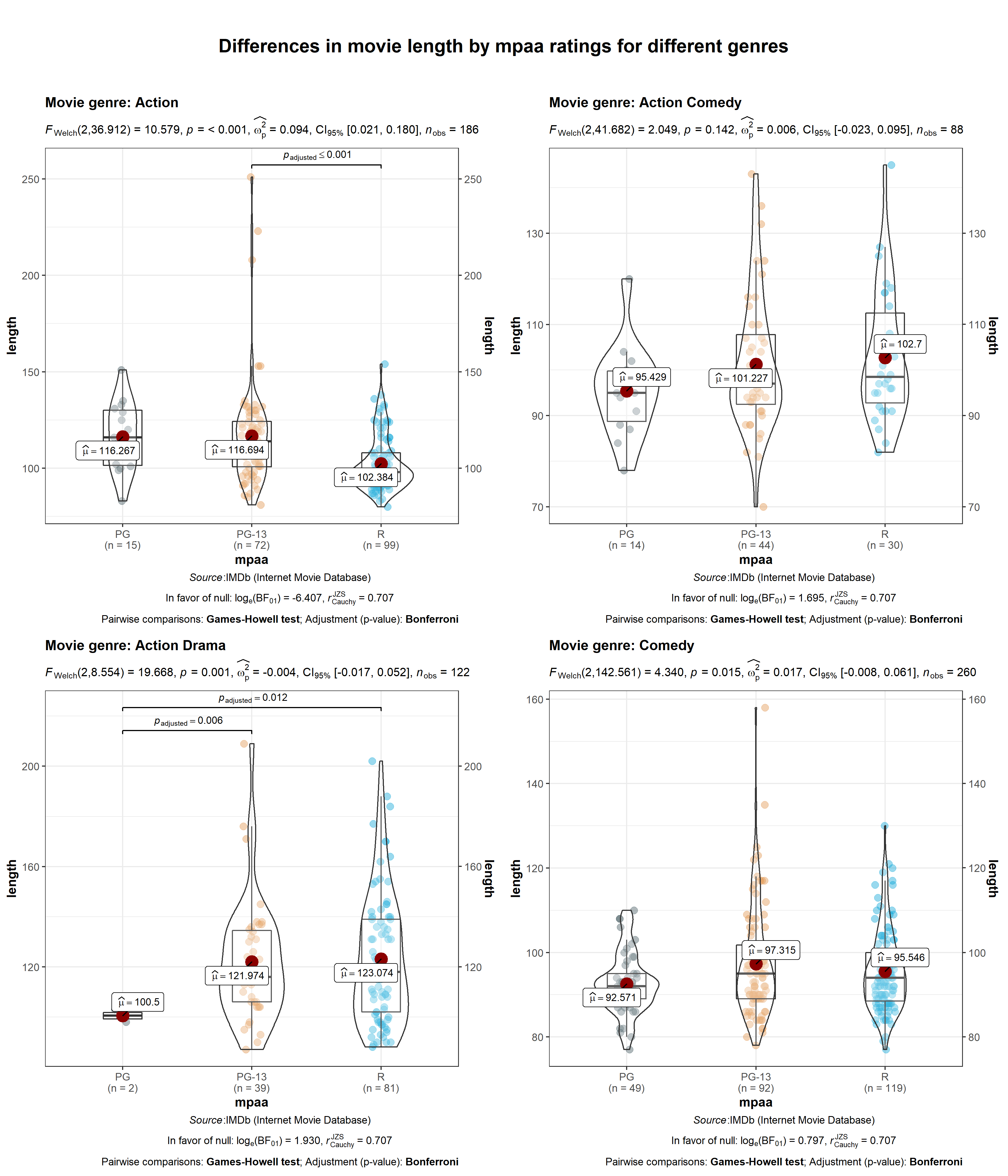
ggstatsplot::grouped_ggbetweenstats(
data = dplyr::filter(
.data = ggstatsplot::movies_long,
genre %in% c("Action", "Action Comedy", "Action Drama", "Comedy")
),
x = mpaa,
y = length,
grouping.var = genre, # grouping variable
pairwise.comparisons = TRUE, # display significant pairwise comparisons
p.adjust.method = "bonferroni", # method for adjusting p-values for multiple comparisons
# adding new components to `ggstatsplot` default
ggplot.component = list(ggplot2::scale_y_continuous(sec.axis = ggplot2::dup_axis())),
k = 3,
title.prefix = "Movie genre",
caption = substitute(paste(italic("Source"), ":IMDb (Internet Movie Database)")),
palette = "default_jama",
package = "ggsci",
messages = FALSE,
plotgrid.args = list(nrow = 2),
title.text = "Differences in movie length by mpaa ratings for different genres"
)
Following (between-subjects) tests are carried out for each type of analyses-
| Type | No. of groups | Test |
|---|---|---|
| Parametric | > 2 | Fisher’s or Welch’s one-way ANOVA |
| Non-parametric | > 2 | Kruskal-Wallis one-way ANOVA |
| Robust | > 2 | Heteroscedastic one-way ANOVA for trimmed means |
| Bayes Factor | > 2 | Fisher’s ANOVA |
| Parametric | 2 | Student’s or Welch’s t-test |
| Non-parametric | 2 | Mann-Whitney U test |
| Robust | 2 | Yuen’s test for trimmed means |
| Bayes Factor | 2 | Student’s t-test |
The omnibus effect in one-way ANOVA design can also be followed up with more focal pairwise comparison tests. Here is a summary of multiple pairwise comparison tests supported in ggbetweenstats-
| Type | Equal variance? | Test | p-value adjustment? |
|---|---|---|---|
| Parametric | No | Games-Howell test | Yes |
| Parametric | Yes | Student’s t-test | Yes |
| Non-parametric | No | Dunn test | Yes |
| Robust | No | Yuen’s trimmed means test | Yes |
| Bayes Factor | NA |
Student’s t-test | NA |
For more, see the ggbetweenstats vignette: https://indrajeetpatil.github.io/ggstatsplot/articles/web_only/ggbetweenstats.html
ggwithinstatsggbetweenstats function has an identical twin function ggwithinstats for repeated measures designs that behaves in the same fashion with a few minor tweaks introduced to properly visualize the repeated measures design. As can be seen from an example below, the only difference between the plot structure is that now the group means are connected by paths to highlight the fact that these data are paired with each other.
# for reproducibility and data
set.seed(123)
library(WRS2)
# plot
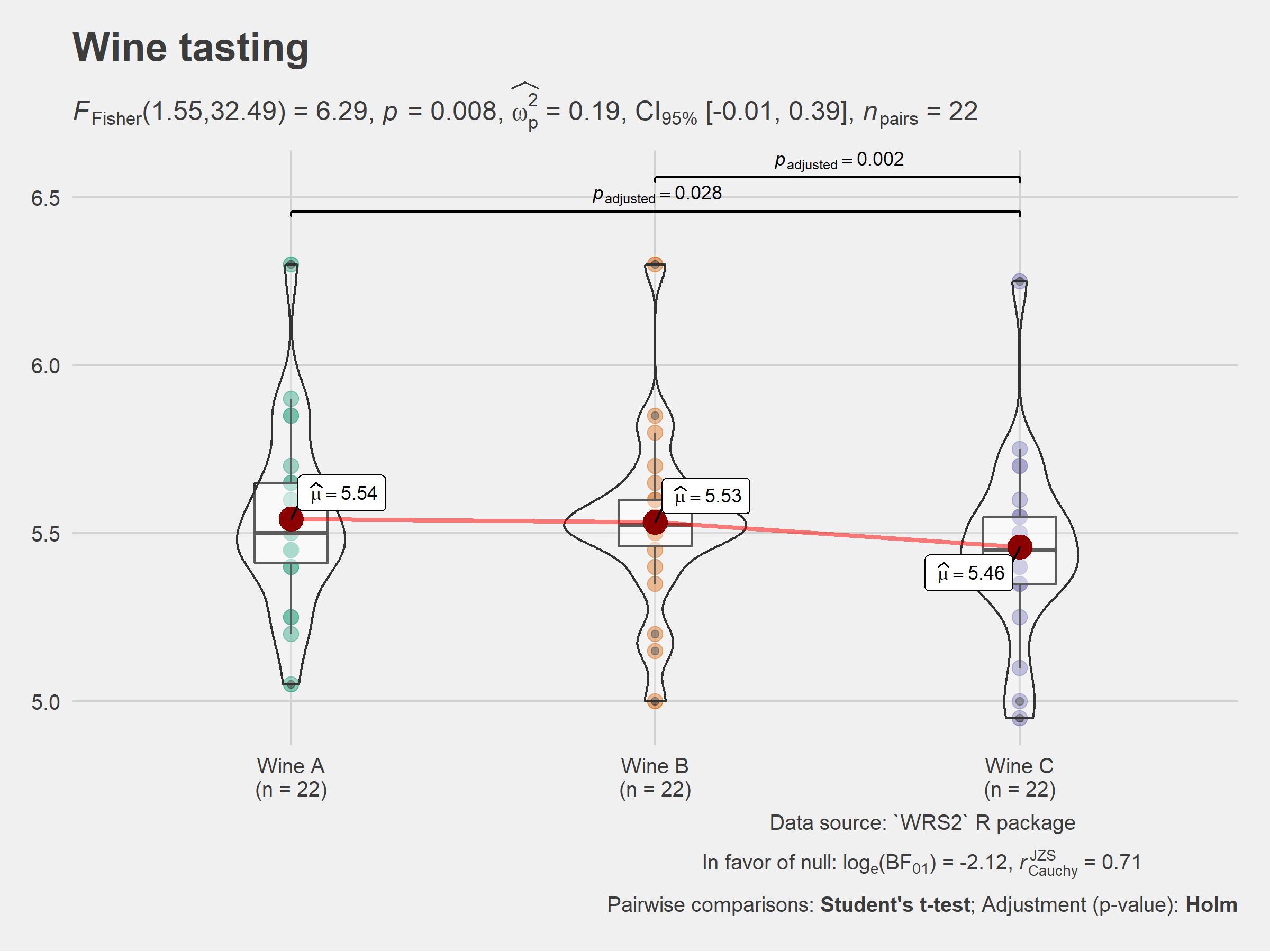
ggstatsplot::ggwithinstats(
data = WineTasting,
x = Wine,
y = Taste,
pairwise.comparisons = TRUE, # show pairwise comparison test results
title = "Wine tasting",
caption = "Data source: `WRS2` R package",
ggtheme = ggthemes::theme_fivethirtyeight(),
ggstatsplot.layer = FALSE,
messages = FALSE
)
As with the ggbetweenstats, this function also has a grouped_ variant that makes repeating the same analysis across a single grouping variable quicker. We will see an example with only repeated measurements-
# common setup
set.seed(123)
# plot
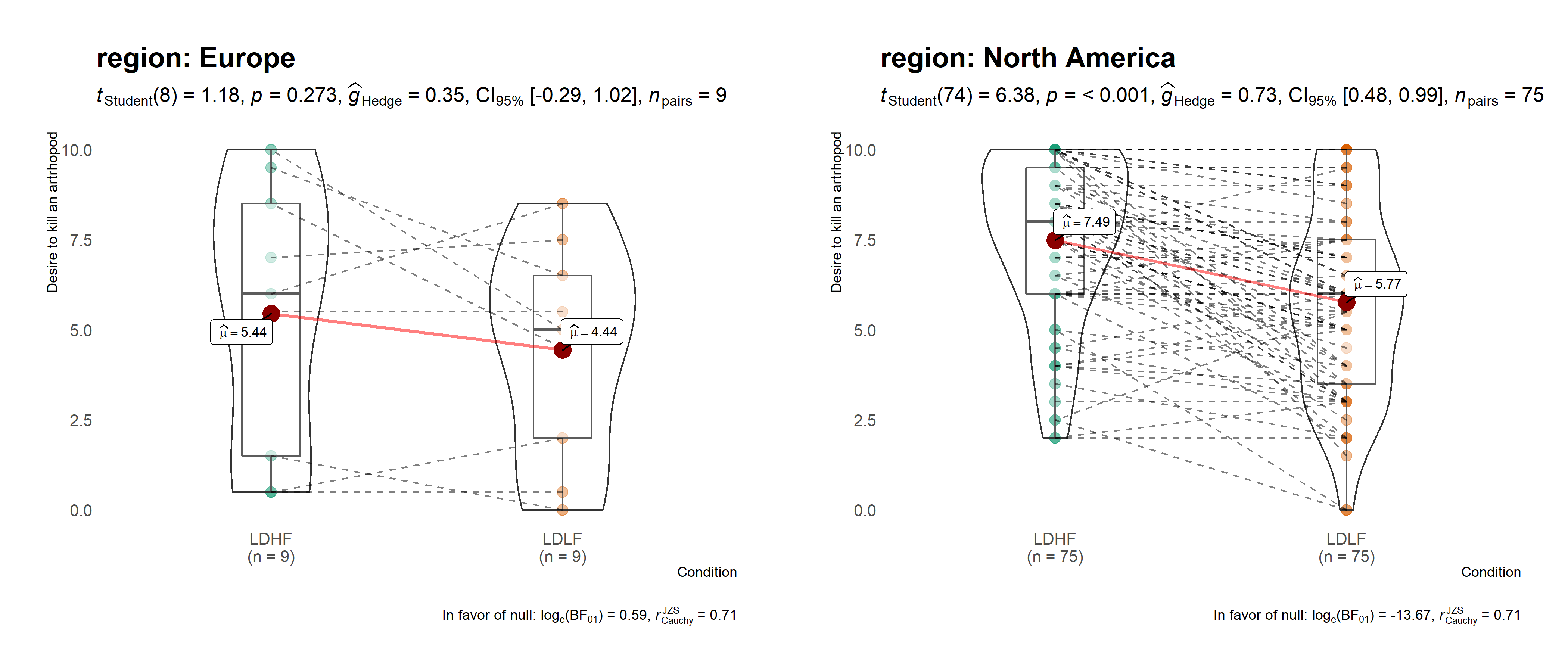
ggstatsplot::grouped_ggwithinstats(
data = dplyr::filter(
.data = ggstatsplot::bugs_long,
region %in% c("Europe", "North America"),
condition %in% c("LDLF", "LDHF")
),
x = condition,
y = desire,
xlab = "Condition",
ylab = "Desire to kill an artrhopod",
grouping.var = region,
outlier.tagging = TRUE,
outlier.label = education,
ggtheme = hrbrthemes::theme_ipsum_tw(),
ggstatsplot.layer = FALSE,
messages = FALSE
)
Following (within-subjects) tests are carried out for each type of analyses-
| Type | No. of groups | Test |
|---|---|---|
| Parametric | > 2 | One-way repeated measures ANOVA |
| Non-parametric | > 2 | Friedman’s rank sum test |
| Robust | > 2 | Heteroscedastic one-way repeated measures ANOVA for trimmed means |
| Bayes Factor | > 2 | One-way repeated measures ANOVA |
| Parametric | 2 | Student’s t-test |
| Non-parametric | 2 | Wilcoxon signed-rank test |
| Robust | 2 | Yuen’s test on trimmed means for dependent samples |
| Bayes Factor | 2 | Student’s t-test |
The omnibus effect in one-way ANOVA design can also be followed up with more focal pairwise comparison tests. Here is a summary of multiple pairwise comparison tests supported in ggwithinstats-
| Type | Test | p-value adjustment? |
|---|---|---|
| Parametric | Student’s t-test | Yes |
| Non-parametric | Durbin-Conover test | Yes |
| Robust | Yuen’s trimmed means test | Yes |
| Bayes Factor | Student’s t-test | NA |
For more, see the ggwithinstats vignette: https://indrajeetpatil.github.io/ggstatsplot/articles/web_only/ggwithinstats.html
ggscatterstatsThis function creates a scatterplot with marginal distributions overlaid on the axes (from ggExtra::ggMarginal) and results from statistical tests in the subtitle:
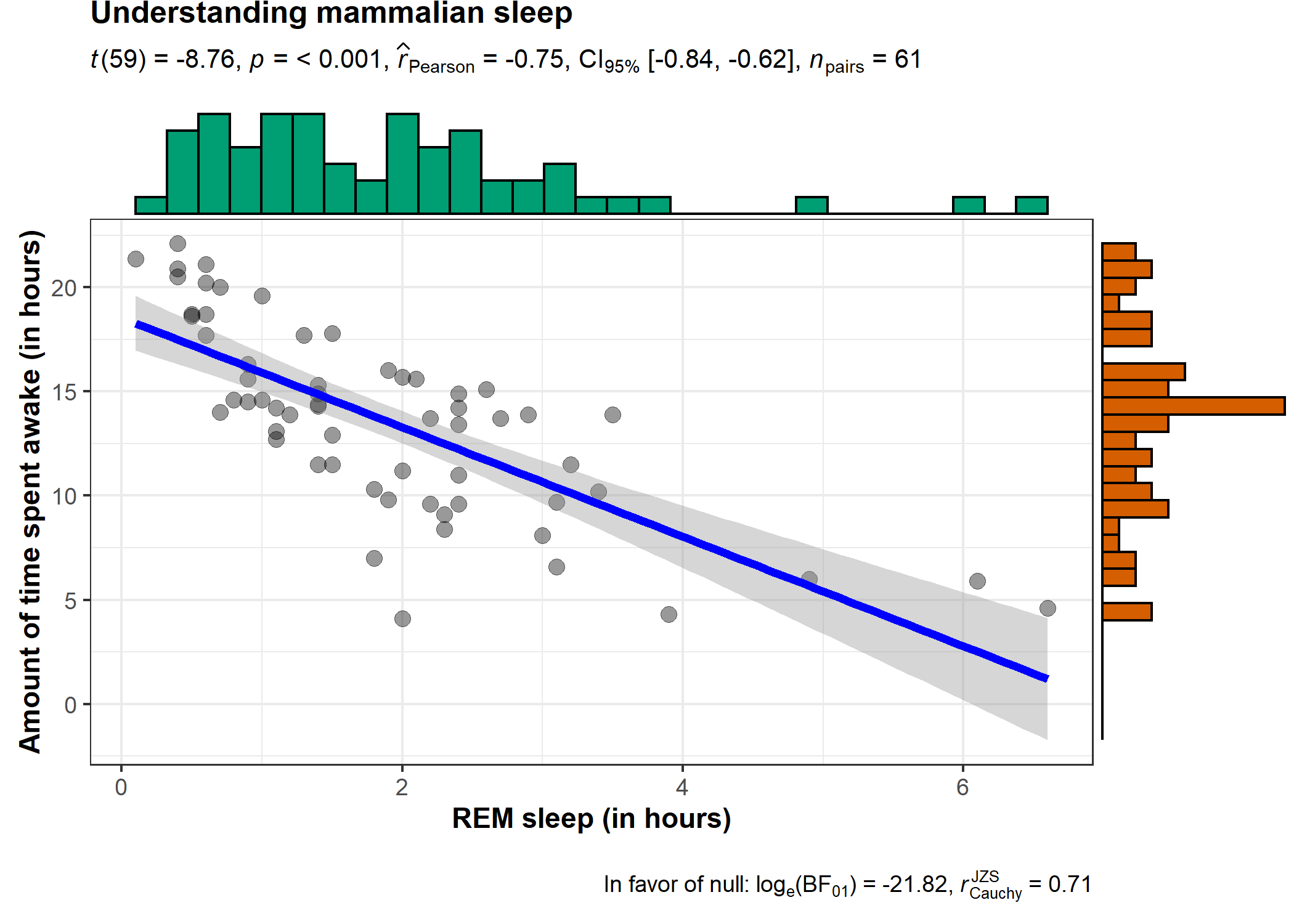
ggstatsplot::ggscatterstats(
data = ggplot2::msleep,
x = sleep_rem,
y = awake,
xlab = "REM sleep (in hours)",
ylab = "Amount of time spent awake (in hours)",
title = "Understanding mammalian sleep",
messages = FALSE
)
The available marginal distributions are-
Number of other arguments can be specified to modify this basic plot-
# for reproducibility
set.seed(123)
# plot
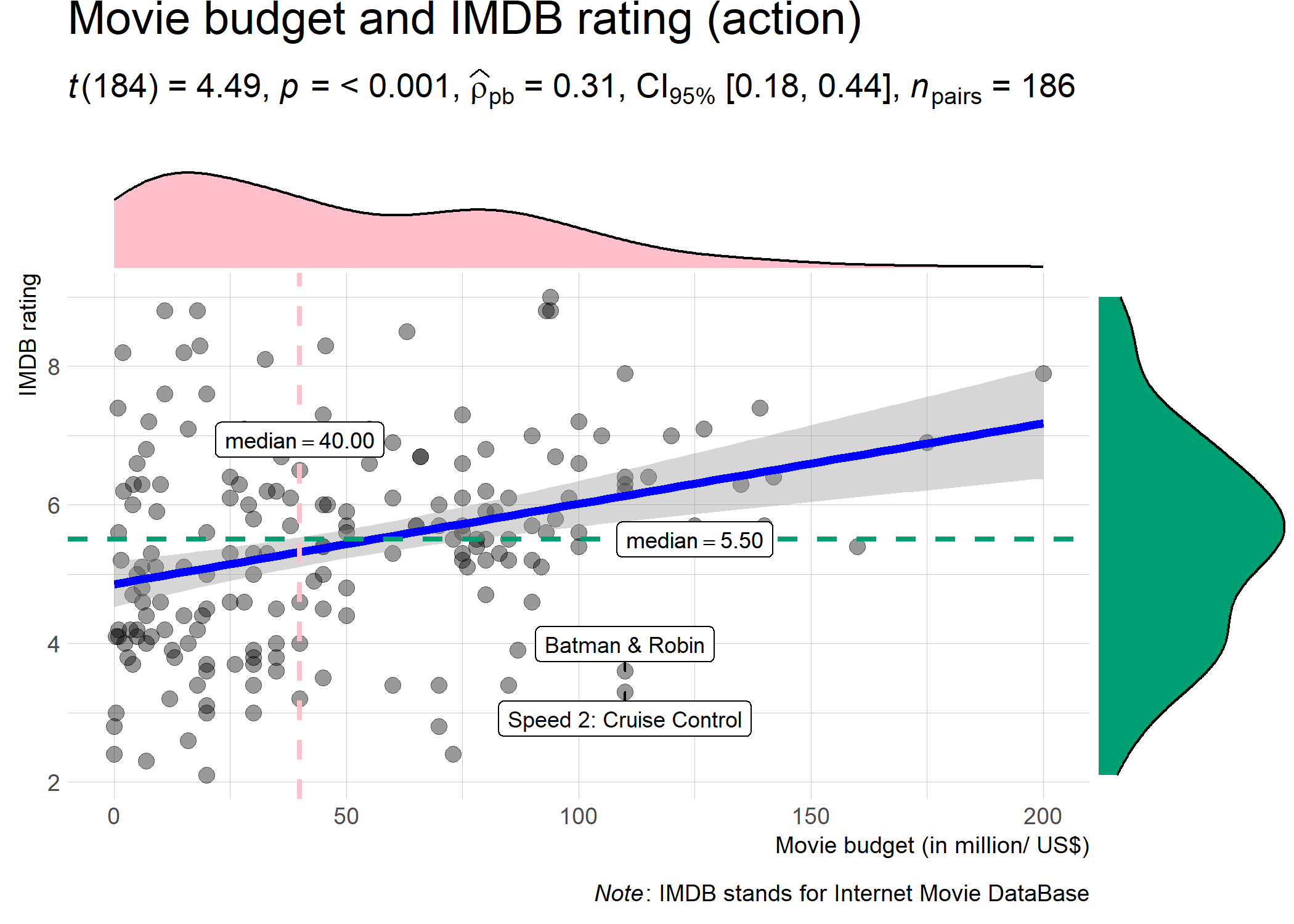
ggstatsplot::ggscatterstats(
data = dplyr::filter(.data = ggstatsplot::movies_long, genre == "Action"),
x = budget,
y = rating,
type = "robust", # type of test that needs to be run
xlab = "Movie budget (in million/ US$)", # label for x axis
ylab = "IMDB rating", # label for y axis
label.var = "title", # variable for labeling data points
label.expression = "rating < 5 & budget > 100", # expression that decides which points to label
title = "Movie budget and IMDB rating (action)", # title text for the plot
caption = expression(paste(italic("Note"), ": IMDB stands for Internet Movie DataBase")),
ggtheme = hrbrthemes::theme_ipsum_ps(), # choosing a different theme
ggstatsplot.layer = FALSE, # turn off `ggstatsplot` theme layer
marginal.type = "density", # type of marginal distribution to be displayed
xfill = "pink", # color fill for x-axis marginal distribution
yfill = "#009E73", # color fill for y-axis marginal distribution
centrality.parameter = "median", # central tendency lines to be displayed
messages = FALSE # turn off messages and notes
)
Additionally, there is also a grouped_ variant of this function that makes it easy to repeat the same operation across a single grouping variable. Also, note that, as opposed to the other functions, this function does not return a ggplot object and any modification you want to make can be made in advance using ggplot.component argument (available for all functions, but especially useful for this particular function):
# for reproducibility
set.seed(123)
# plot
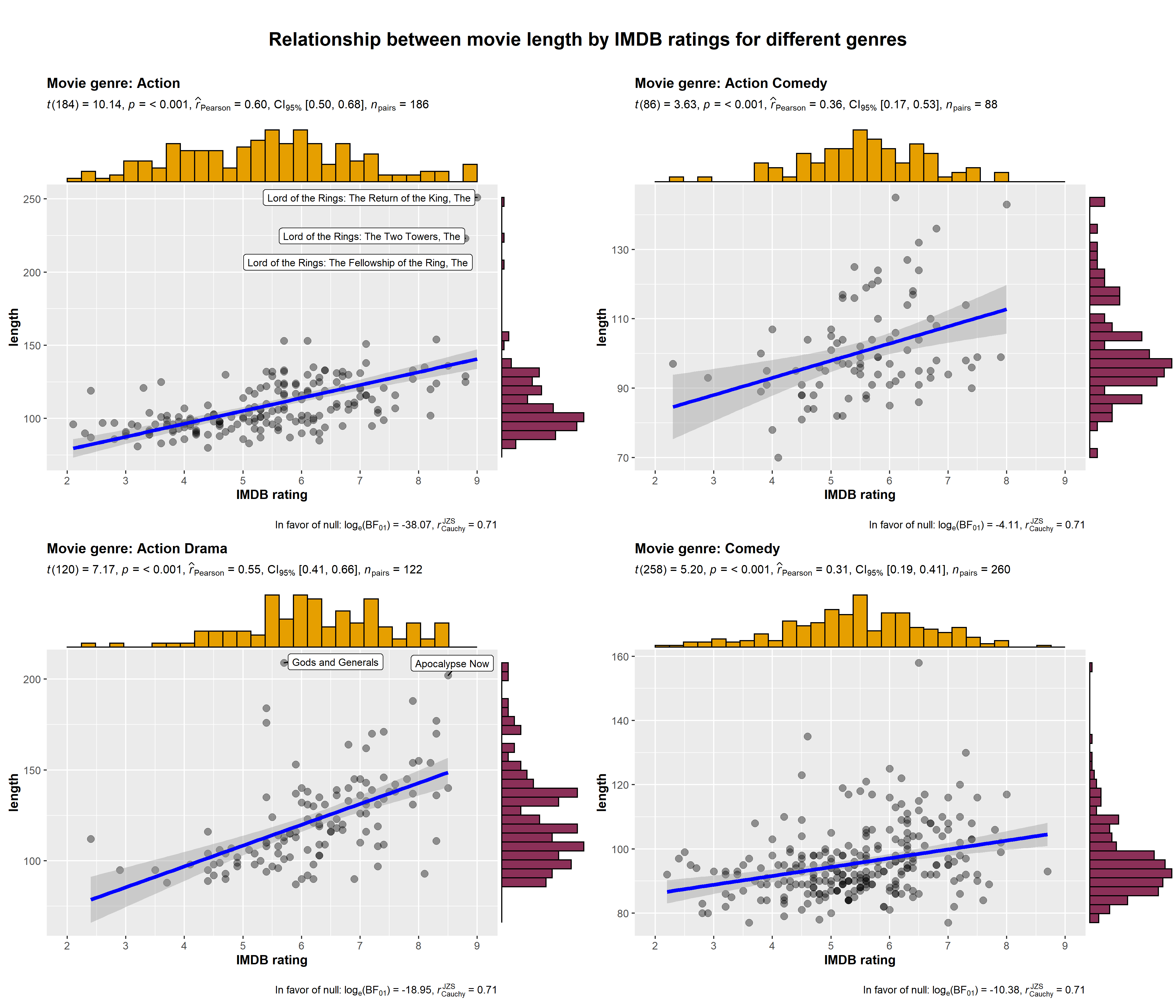
ggstatsplot::grouped_ggscatterstats(
data = dplyr::filter(
.data = ggstatsplot::movies_long,
genre %in% c("Action", "Action Comedy", "Action Drama", "Comedy")
),
x = rating,
y = length,
grouping.var = genre, # grouping variable
label.var = title,
label.expression = length > 200,
xfill = "#E69F00",
yfill = "#8b3058",
xlab = "IMDB rating",
title.prefix = "Movie genre",
ggtheme = ggplot2::theme_grey(),
ggplot.component = list(
ggplot2::scale_x_continuous(breaks = seq(2, 9, 1), limits = (c(2, 9)))
),
messages = FALSE,
plotgrid.args = list(nrow = 2),
title.text = "Relationship between movie length by IMDB ratings for different genres"
)
Following tests are carried out for each type of analyses. Additionally, the correlation coefficients (and their confidence intervals) are used as effect sizes-
| Type | Test | CI? |
|---|---|---|
| Parametric | Pearson’s correlation coefficient | Yes |
| Non-parametric | Spearman’s rank correlation coefficient | Yes |
| Robust | Percentage bend correlation coefficient | Yes |
| Bayes Factor | Pearson’s correlation coefficient | No |
For more, see the ggscatterstats vignette: https://indrajeetpatil.github.io/ggstatsplot/articles/web_only/ggscatterstats.html
ggpiestatsThis function creates a pie chart for categorical or nominal variables with results from contingency table analysis (Pearson’s chi-squared test for between-subjects design and McNemar’s chi-squared test for within-subjects design) included in the subtitle of the plot. If only one categorical variable is entered, results from one-sample proportion test (i.e., a chi-squared goodness of fit test) will be displayed as a subtitle.
To study an interaction between two categorical variables:
# for reproducibility
set.seed(123)
# plot
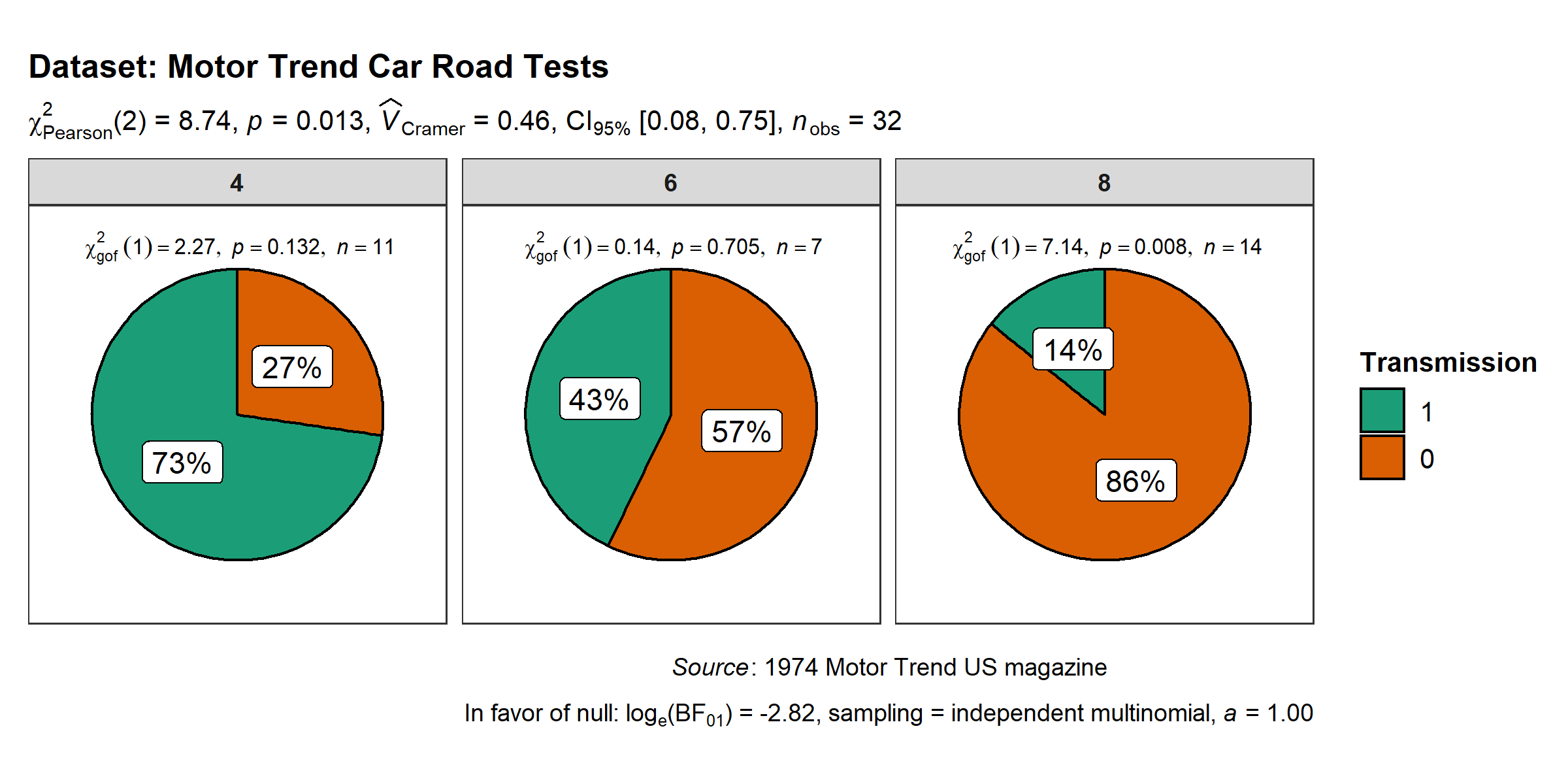
ggstatsplot::ggpiestats(
data = mtcars,
x = am,
y = cyl,
title = "Dataset: Motor Trend Car Road Tests", # title for the plot
legend.title = "Transmission", # title for the legend
caption = substitute(paste(italic("Source"), ": 1974 Motor Trend US magazine")),
messages = FALSE
)
In case of repeated measures designs, setting paired = TRUE will produce results from McNemar’s chi-squared test-
# for reproducibility
set.seed(123)
# plot
ggstatsplot::ggpiestats(
data = data.frame(
"before" = c("Approve", "Approve", "Disapprove", "Disapprove"),
"after" = c("Approve", "Disapprove", "Approve", "Disapprove"),
counts = c(794, 150, 86, 570),
check.names = FALSE
),
x = before,
y = after,
counts = counts,
title = "Survey results before and after the intervention",
label = "both",
paired = TRUE, # within-subjects design
package = "wesanderson",
palette = "Royal1"
)
#> Note: 95% CI for effect size estimate was computed with 100 bootstrap samples.
#> # A tibble: 2 x 11
#> after counts perc N Approve Disapprove statistic p.value
#> <fct> <int> <dbl> <chr> <chr> <chr> <dbl> <dbl>
#> 1 Disapprove 720 45 (n = 720) 20.83% 79.17% 245 3.20e- 55
#> 2 Approve 880 55. (n = 880) 90.23% 9.77% 570. 6.80e-126
#> parameter method significance
#> <dbl> <chr> <chr>
#> 1 1 Chi-squared test for given probabilities ***
#> 2 1 Chi-squared test for given probabilities ***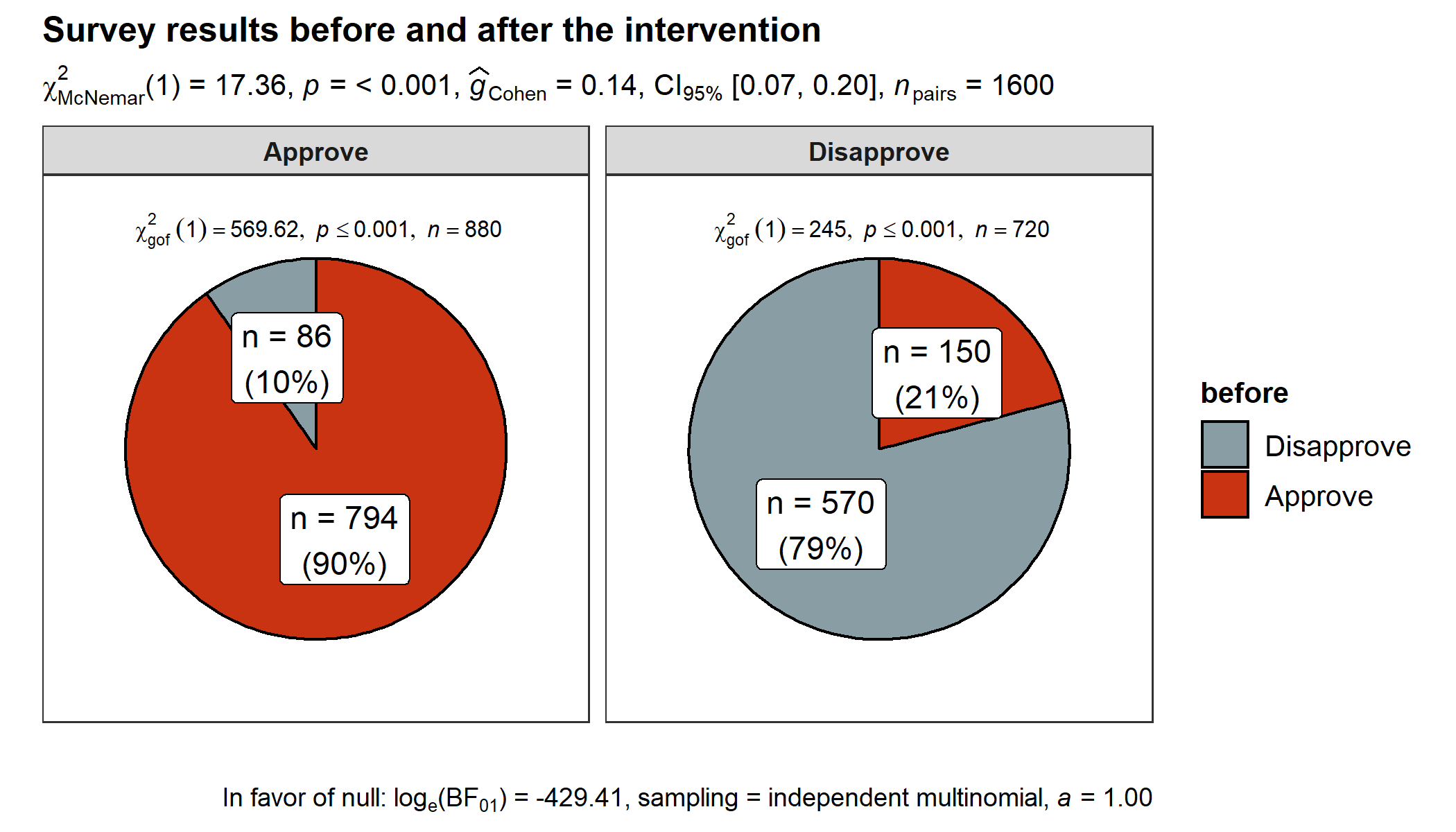
Additionally, there is also a grouped_ variant of this function that makes it easy to repeat the same operation across a single grouping variable. Following example is a case where the theoretical question is about proportions for different levels of a single nominal variable:
# for reproducibility
set.seed(123)
# plot
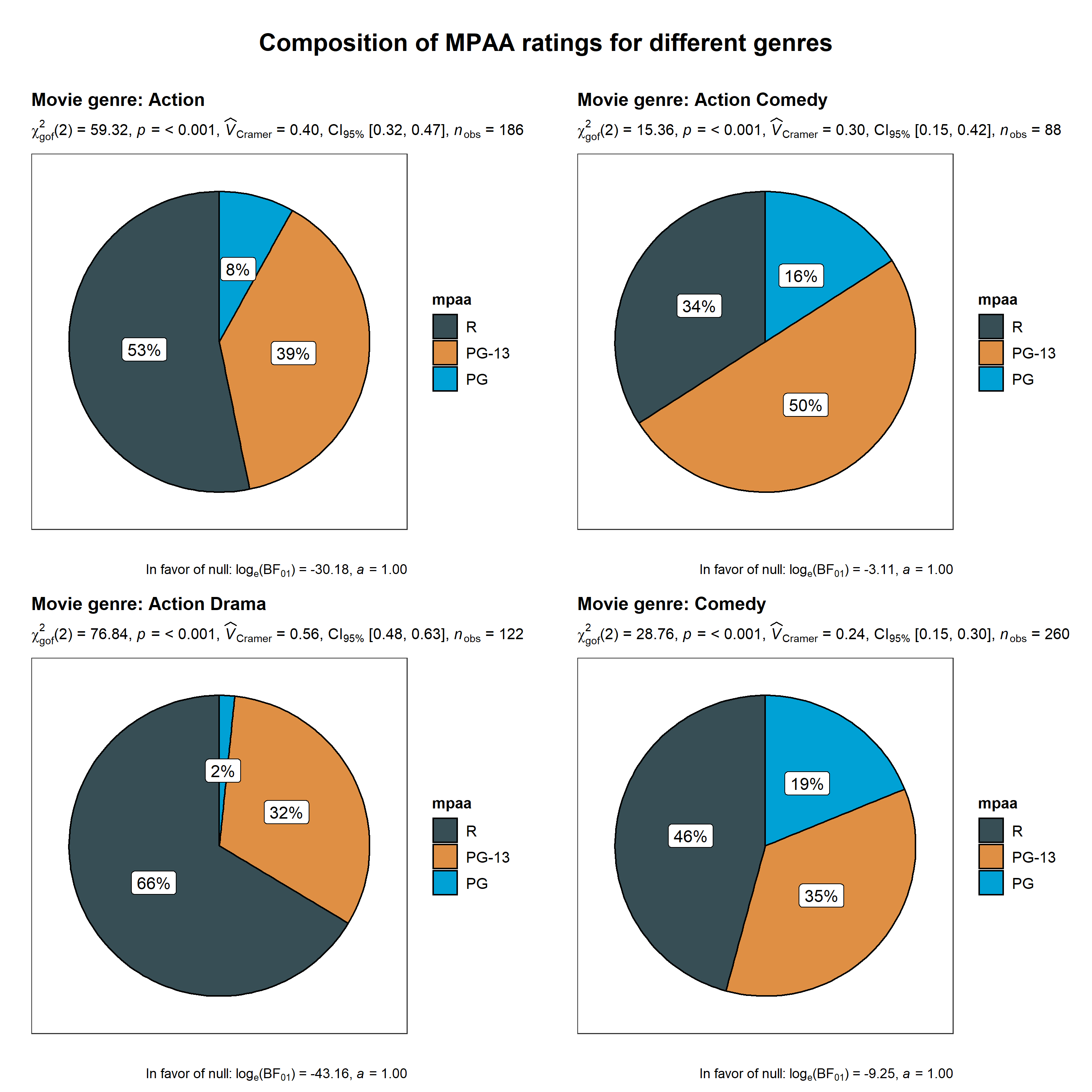
ggstatsplot::grouped_ggpiestats(
dplyr::filter(
.data = ggstatsplot::movies_long,
genre %in% c("Action", "Action Comedy", "Action Drama", "Comedy")
),
x = mpaa,
grouping.var = genre, # grouping variable
title.prefix = "Movie genre", # prefix for the facetted title
messages = FALSE,
package = "ggsci", # package from which color palette is to be taken
palette = "default_jama", # choosing a different color palette
title.text = "Composition of MPAA ratings for different genres"
)
Following tests are carried out for each type of analyses-
| Type of data | Design | Test |
|---|---|---|
| Unpaired | Pearson’s |
|
| Paired | McNemar’s |
|
| Frequency | Goodness of fit ( |
Following effect sizes (and confidence intervals/CI) are available for each type of test-
| Type | Effect size | CI? |
|---|---|---|
| Pearson’s chi-squared test | Cramér’s V | Yes |
| McNemar’s test | Cohen’s g | Yes |
| Goodness of fit | Cramér’s V | Yes |
For more, see the ggpiestats vignette: https://indrajeetpatil.github.io/ggstatsplot/articles/web_only/ggpiestats.html
ggbarstatsIn case you are not a fan of pie charts (for very good reasons), you can alternatively use ggbarstats function which has a similar syntax-
# for reproducibility
set.seed(123)
library(ggplot2)
# plot
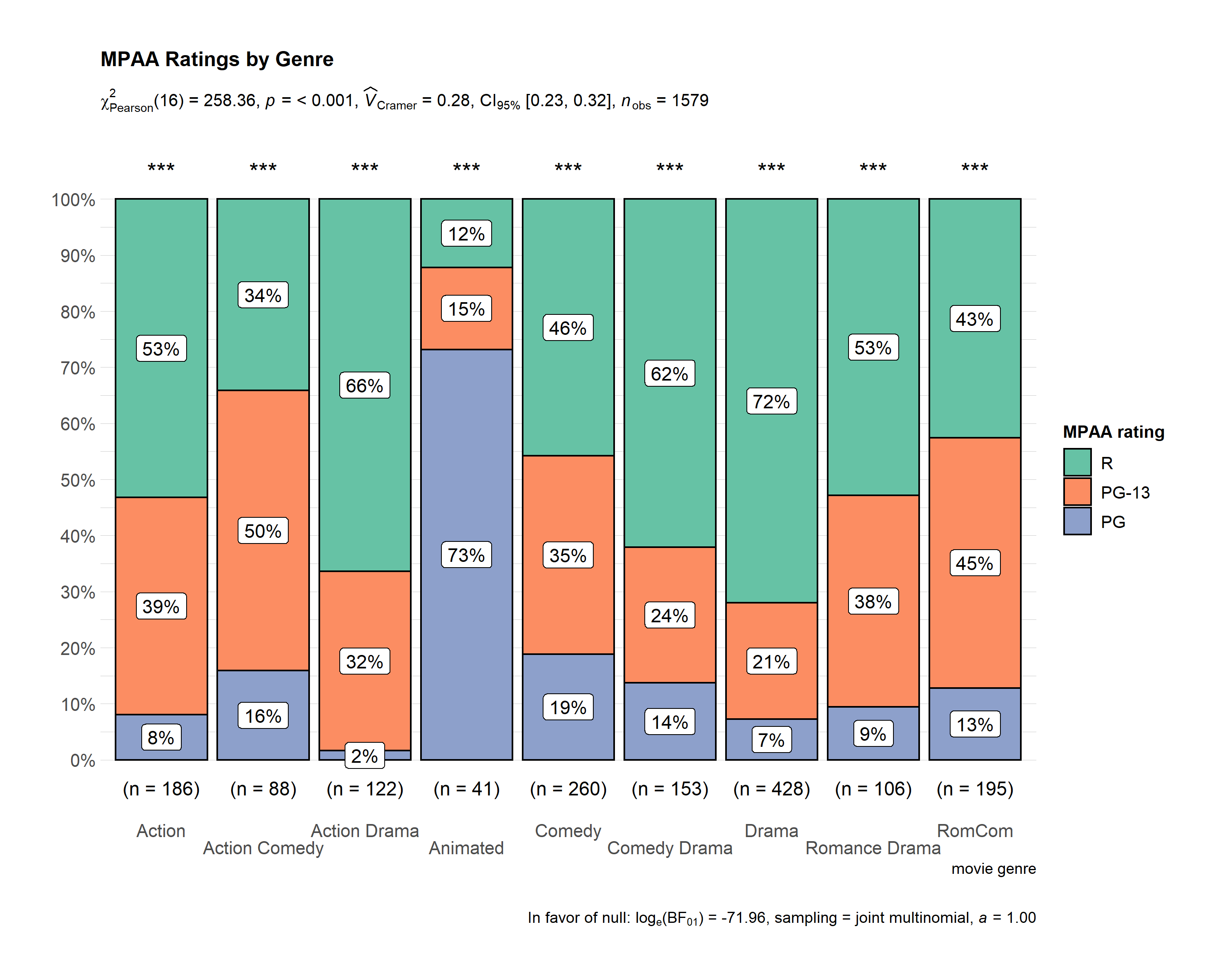
ggstatsplot::ggbarstats(
data = ggstatsplot::movies_long,
x = mpaa,
y = genre,
sampling.plan = "jointMulti",
title = "MPAA Ratings by Genre",
xlab = "movie genre",
legend.title = "MPAA rating",
ggtheme = hrbrthemes::theme_ipsum_pub(),
ggplot.component = list(scale_x_discrete(guide = guide_axis(n.dodge = 2))),
palette = "Set2",
messages = FALSE
)
And, needless to say, there is also a grouped_ variant of this function-
# setup
set.seed(123)
# smaller dataset
df <-
dplyr::filter(
.data = forcats::gss_cat,
race %in% c("Black", "White"),
relig %in% c("Protestant", "Catholic", "None"),
!partyid %in% c("No answer", "Don't know", "Other party")
)
# plot
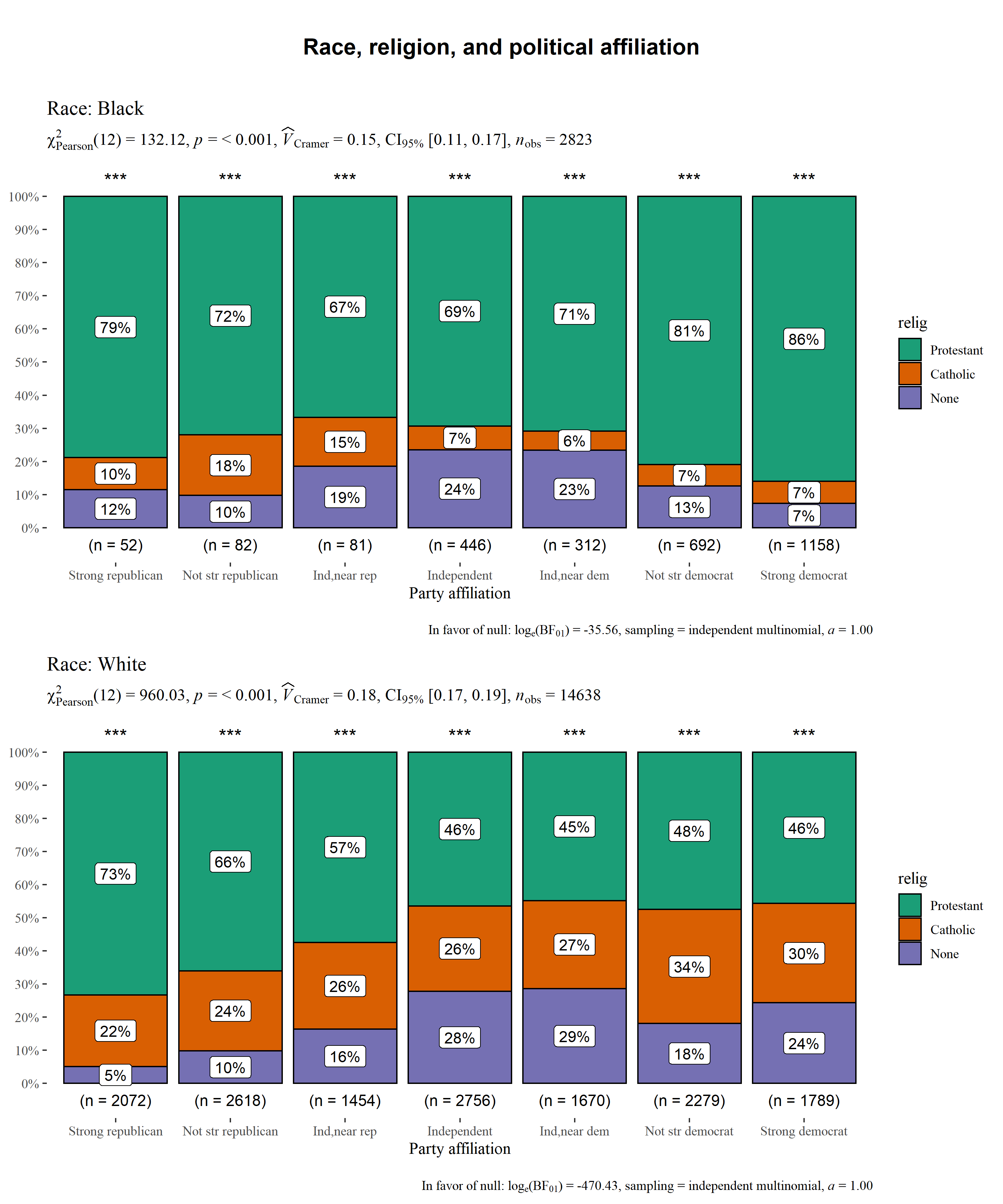
ggstatsplot::grouped_ggbarstats(
data = df,
x = relig,
y = partyid,
grouping.var = race,
title.prefix = "Race",
xlab = "Party affiliation",
ggtheme = ggthemes::theme_tufte(base_size = 12),
ggstatsplot.layer = FALSE,
messages = FALSE,
title.text = "Race, religion, and political affiliation",
plotgrid.args = list(nrow = 2)
)
This is identical to the ggpiestats function summary of tests.
gghistostatsTo visualize the distribution of a single variable and check if its mean is significantly different from a specified value with a one-sample test, gghistostats can be used.
# for reproducibility
set.seed(123)
# plot
ggstatsplot::gghistostats(
data = iris, # dataframe from which variable is to be taken
x = Sepal.Length, # numeric variable whose distribution is of interest
title = "Distribution of Iris sepal length", # title for the plot
caption = substitute(paste(italic("Source:"), "Ronald Fisher's Iris data set")),
bar.measure = "both",
test.value = 5, # default value is 0
test.value.line = TRUE, # display a vertical line at test value
centrality.parameter = "mean", # which measure of central tendency is to be plotted
centrality.line.args = list(color = "darkred"), # aesthetics for central tendency line
binwidth = 0.10, # binwidth value (experiment)
messages = FALSE, # turn off the messages
ggtheme = hrbrthemes::theme_ipsum_tw(), # choosing a different theme
ggstatsplot.layer = FALSE # turn off ggstatsplot theme layer
)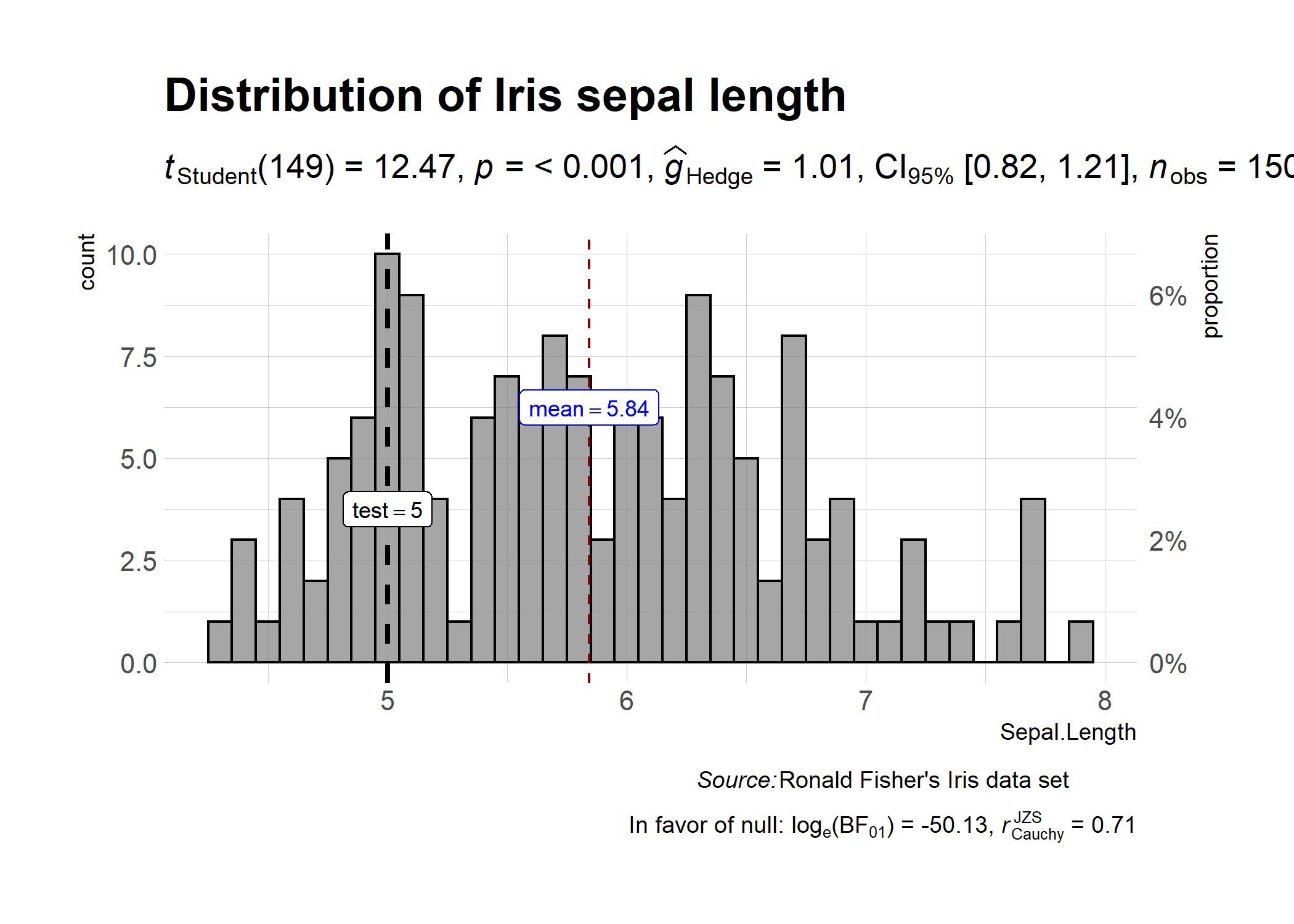
As can be seen from the plot, Bayes Factor can be attached (bf.message = TRUE) to assess evidence in favor of the null hypothesis.
Additionally, there is also a grouped_ variant of this function that makes it easy to repeat the same operation across a single grouping variable:
# for reproducibility
set.seed(123)
# plot
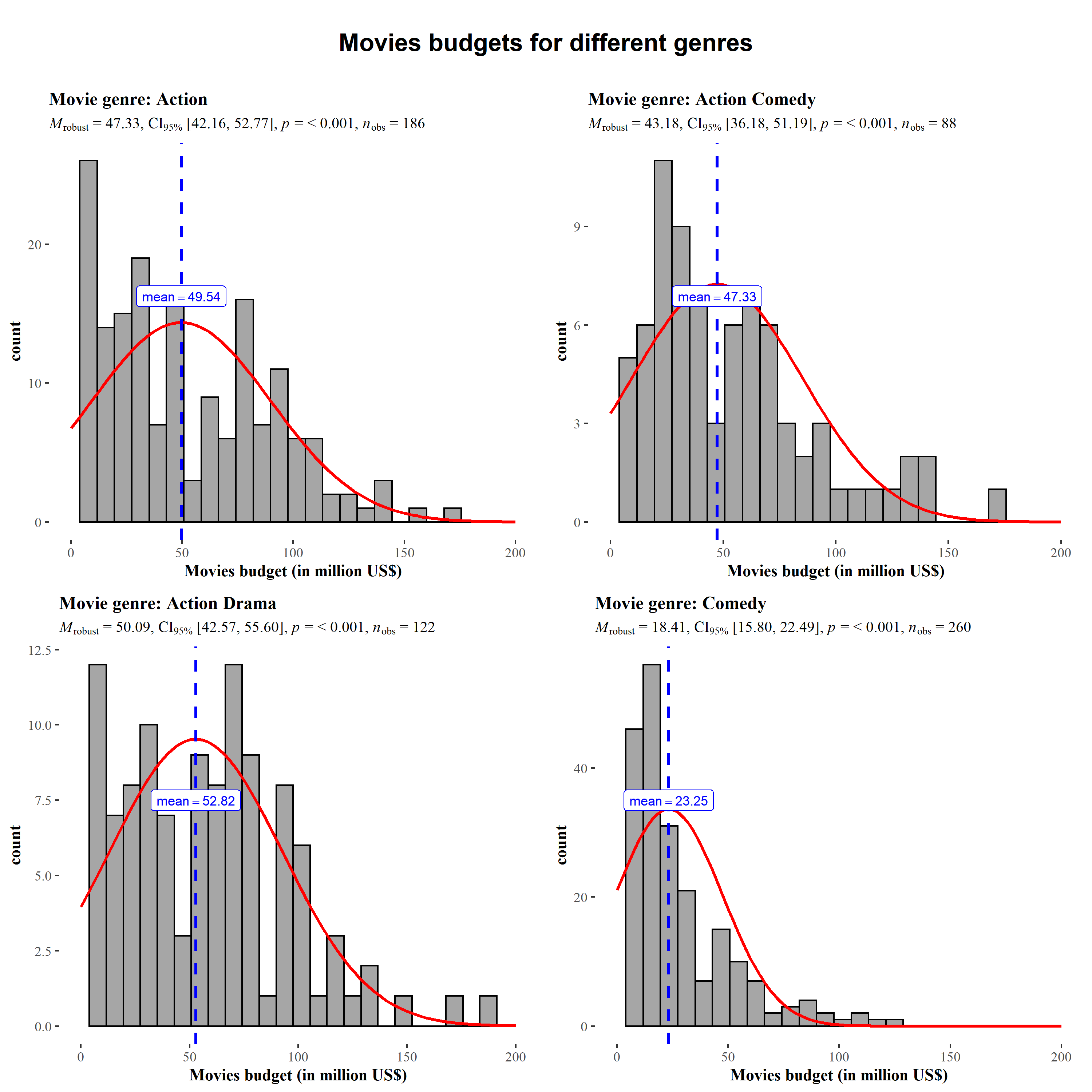
ggstatsplot::grouped_gghistostats(
data = dplyr::filter(
.data = ggstatsplot::movies_long,
genre %in% c("Action", "Action Comedy", "Action Drama", "Comedy")
),
x = budget,
xlab = "Movies budget (in million US$)",
type = "robust", # use robust location measure
grouping.var = genre, # grouping variable
normal.curve = TRUE, # superimpose a normal distribution curve
normal.curve.args = list(color = "red", size = 1),
title.prefix = "Movie genre",
ggtheme = ggthemes::theme_tufte(),
ggplot.component = list( # modify the defaults from `ggstatsplot` for each plot
ggplot2::scale_x_continuous(breaks = seq(0, 200, 50), limits = (c(0, 200)))
),
messages = FALSE,
plotgrid.args = list(nrow = 2),
title.text = "Movies budgets for different genres"
)
Following tests are carried out for each type of analyses-
| Type | Test |
|---|---|
| Parametric | One-sample Student’s t-test |
| Non-parametric | One-sample Wilcoxon test |
| Robust | One-sample percentile bootstrap |
| Bayes Factor | One-sample Student’s t-test |
Following effect sizes (and confidence intervals/CI) are available for each type of test-
| Type | Effect size | CI? |
|---|---|---|
| Parametric | Cohen’s d, Hedge’s g (central-and noncentral-t distribution based) | Yes |
| Non-parametric | r | Yes |
| Robust | robust location measure | Yes |
| Bayes Factor | No | No |
For more, including information about the variant of this function grouped_gghistostats, see the gghistostats vignette: https://indrajeetpatil.github.io/ggstatsplot/articles/web_only/gghistostats.html
ggdotplotstatsThis function is similar to gghistostats, but is intended to be used when the numeric variable also has a label.
# for reproducibility
set.seed(123)
# plot
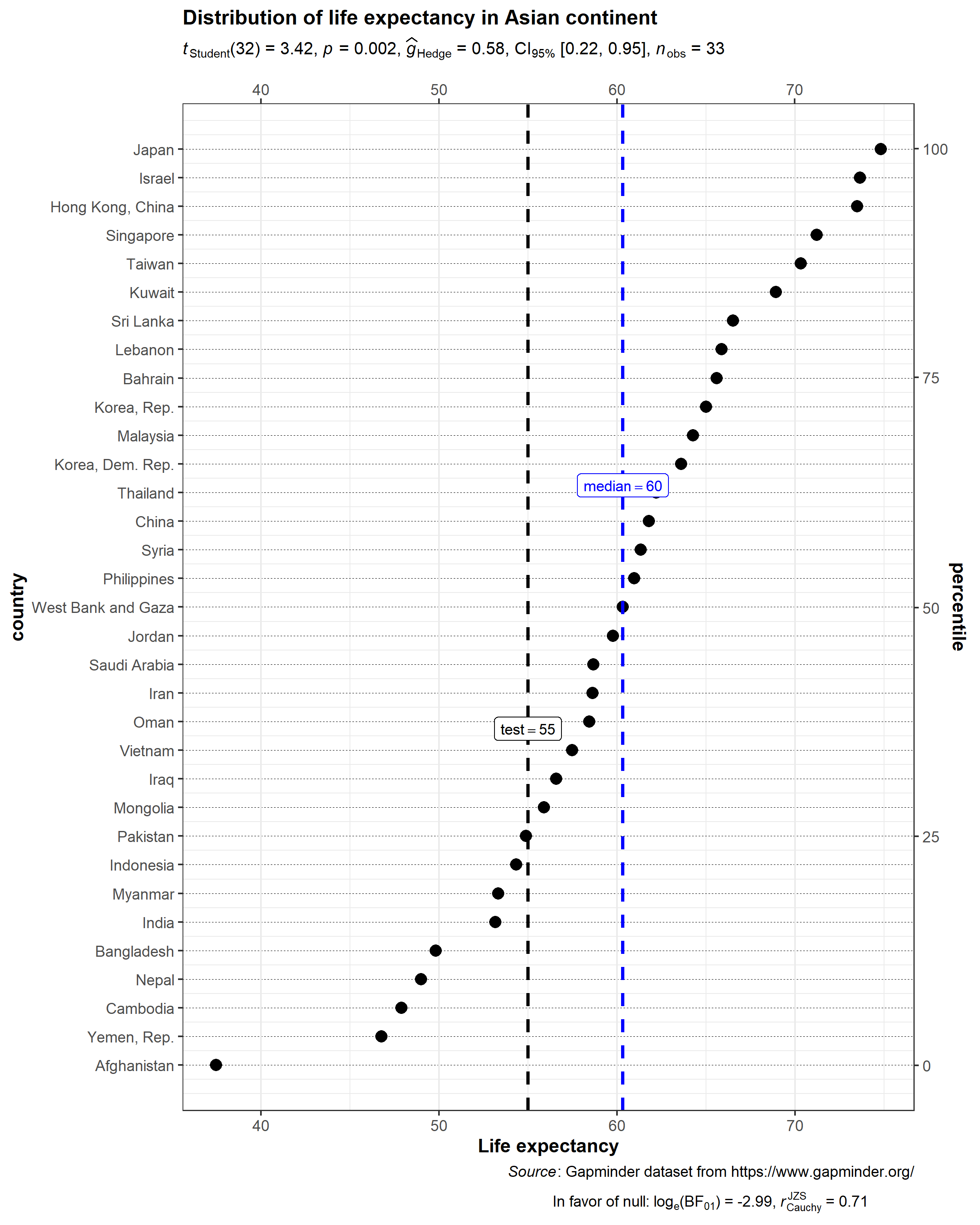
ggdotplotstats(
data = dplyr::filter(.data = gapminder::gapminder, continent == "Asia"),
y = country,
x = lifeExp,
test.value = 55,
test.value.line = TRUE,
centrality.parameter = "median",
centrality.k = 0,
title = "Distribution of life expectancy in Asian continent",
xlab = "Life expectancy",
messages = FALSE,
caption = substitute(
paste(
italic("Source"),
": Gapminder dataset from https://www.gapminder.org/"
)
)
)
As with the rest of the functions in this package, there is also a grouped_ variant of this function to facilitate looping the same operation for all levels of a single grouping variable.
# for reproducibility
set.seed(123)
# plot
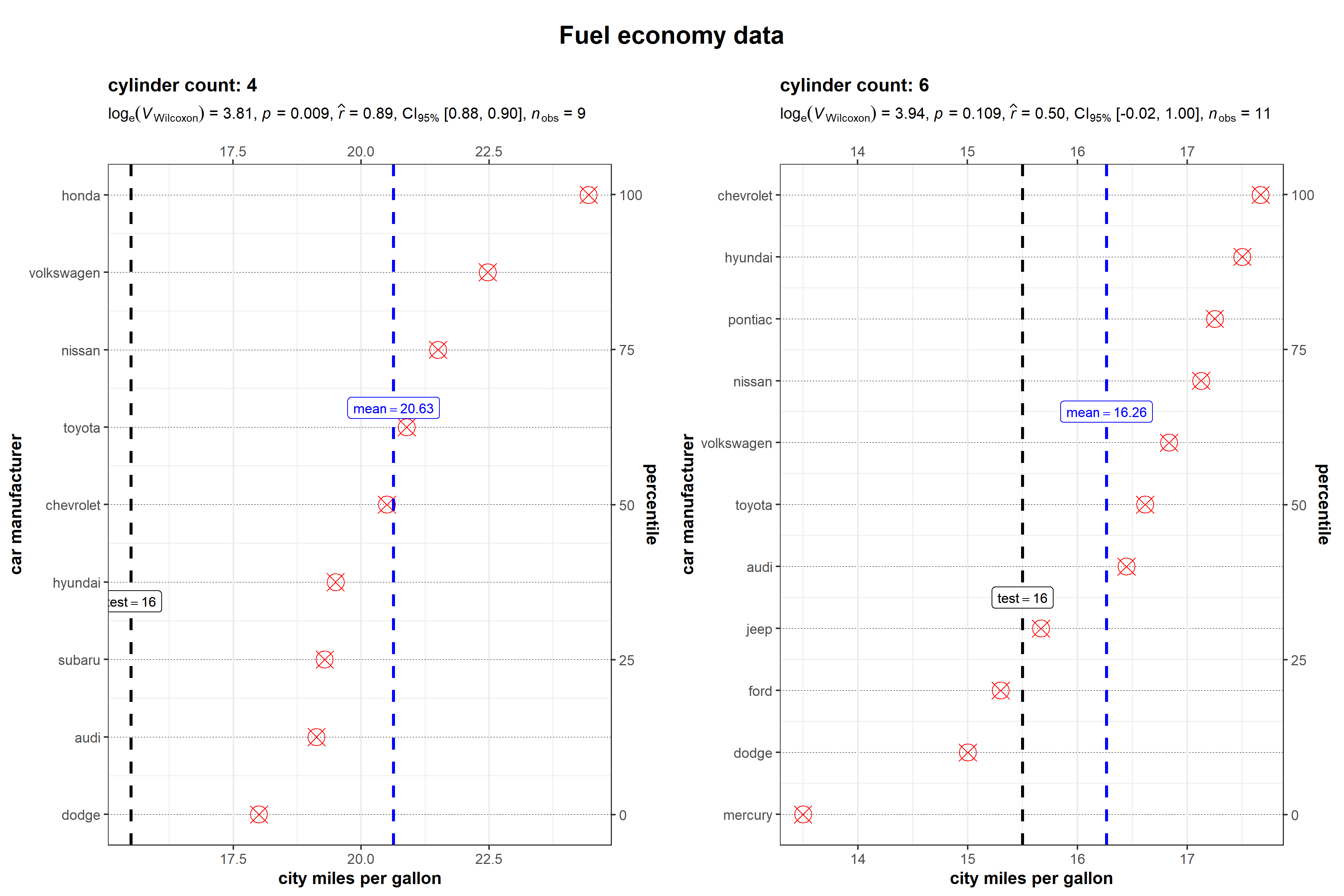
ggstatsplot::grouped_ggdotplotstats(
data = dplyr::filter(.data = ggplot2::mpg, cyl %in% c("4", "6")),
x = cty,
y = manufacturer,
xlab = "city miles per gallon",
ylab = "car manufacturer",
type = "nonparametric", # non-parametric test
grouping.var = cyl, # grouping variable
test.value = 15.5,
test.value.line = TRUE,
title.prefix = "cylinder count",
point.args = list(color = "red", size = 5, shape = 13),
messages = FALSE,
title.text = "Fuel economy data"
)
This is identical to summary of tests for gghistostats.
ggcorrmatggcorrmat makes a correlalogram (a matrix of correlation coefficients) with minimal amount of code. Just sticking to the defaults itself produces publication-ready correlation matrices. But, for the sake of exploring the available options, let’s change some of the defaults. For example, multiple aesthetics-related arguments can be modified to change the appearance of the correlation matrix.
# for reproducibility
set.seed(123)
# as a default this function outputs a correlation matrix plot
ggstatsplot::ggcorrmat(
data = ggplot2::msleep,
type = "robust", # correlation method
p.adjust.method = "holm", # p-value adjustment method for multiple comparisons
cor.vars = c(sleep_rem, awake:bodywt), # a range of variables can be selected
cor.vars.names = c(
"REM sleep", # variable names
"time awake",
"brain weight",
"body weight"
),
matrix.type = "upper", # type of visualization matrix
colors = c("#B2182B", "white", "#4D4D4D"),
title = "Correlalogram for mammals sleep dataset",
subtitle = "sleep units: hours; weight units: kilograms",
caption = "Source: `ggplot2` R package"
)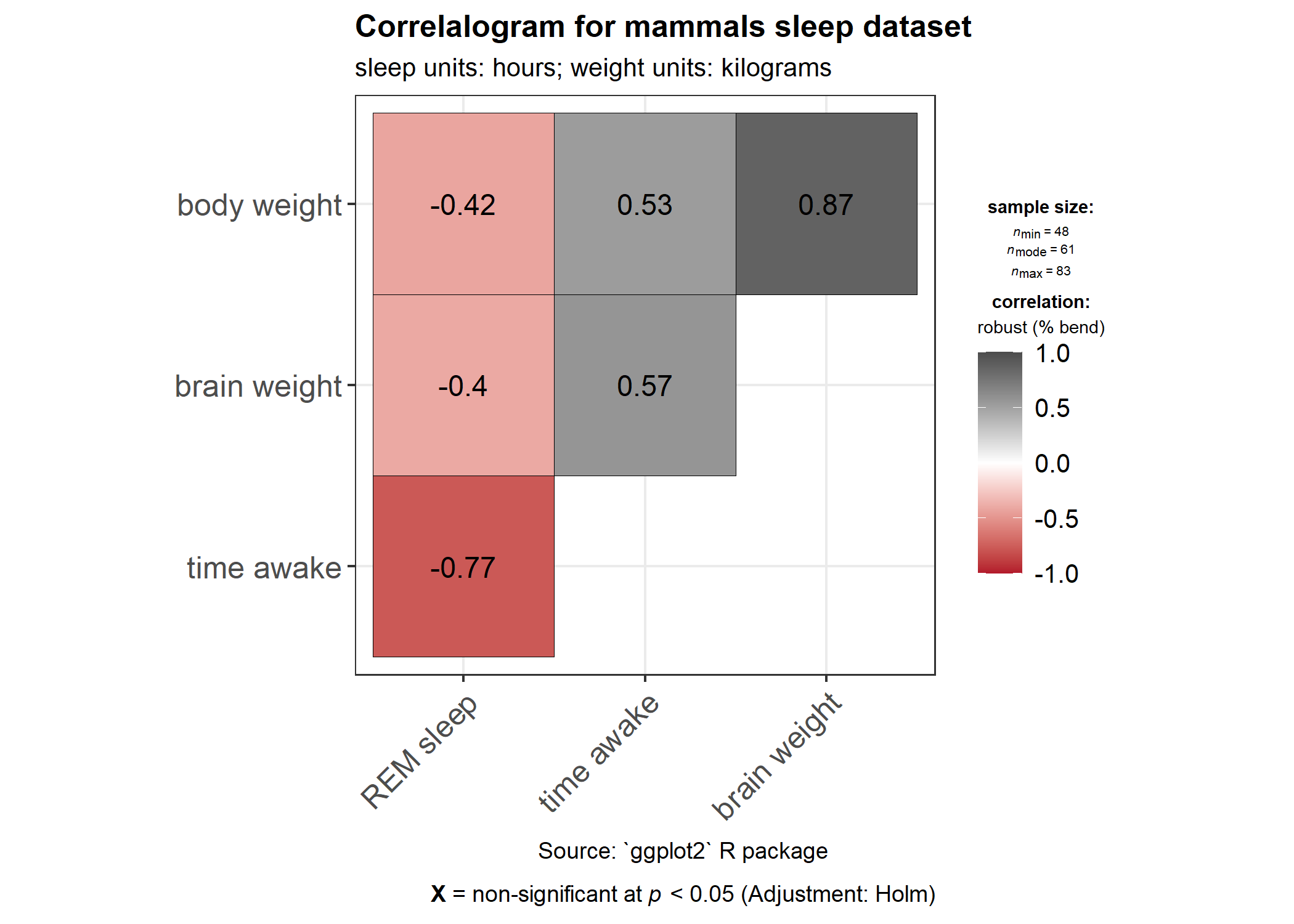
Two things to note:
If there are NAs present in the selected variables, the legend will display minimum, median, and maximum number of pairs used for correlation tests.
If cor.vars are not specified, all numeric variables will be used.
There is also a grouped_ variant of this function that makes it easy to repeat the same operation across a single grouping variable:
# for reproducibility
set.seed(123)
# plot
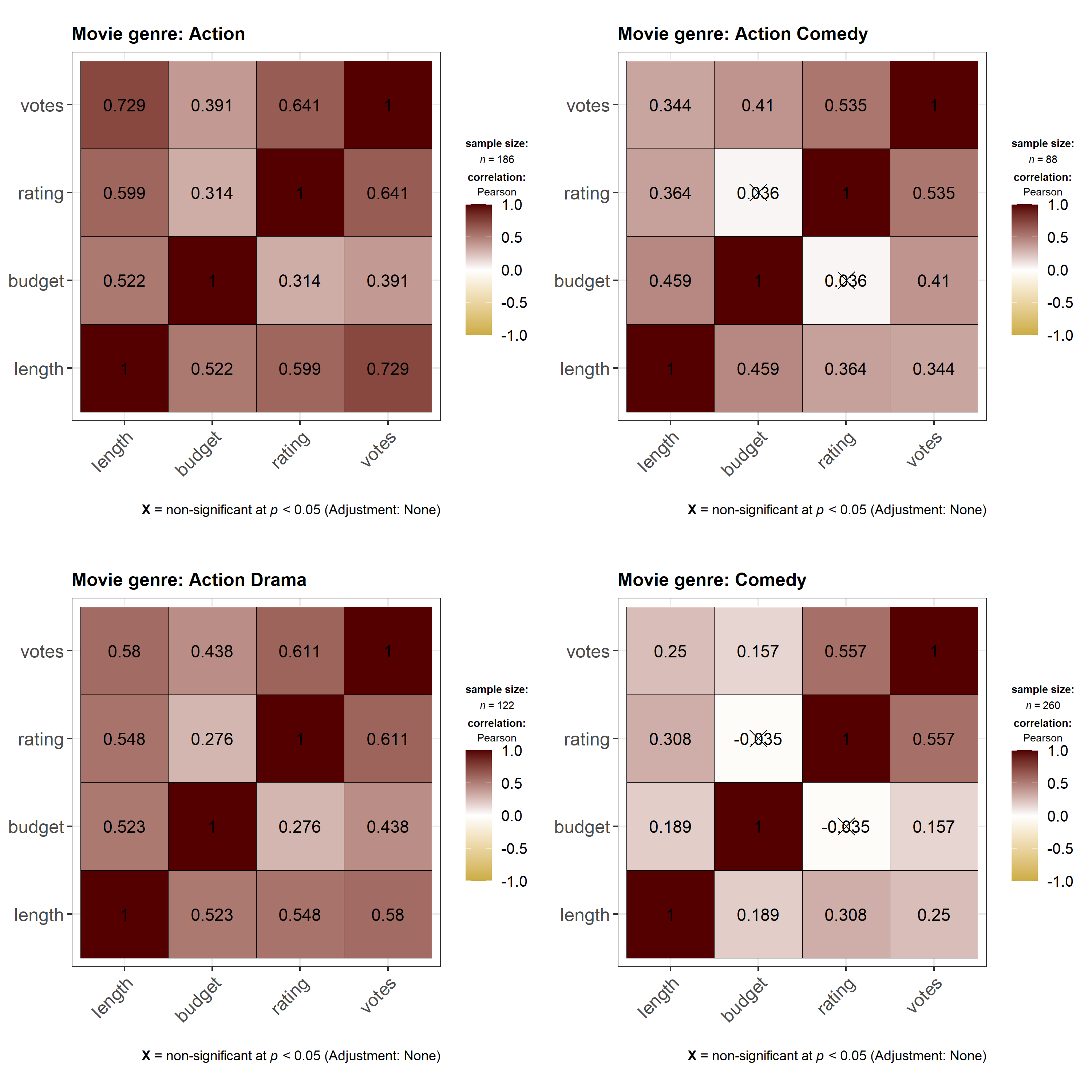
ggstatsplot::grouped_ggcorrmat(
data = dplyr::filter(
.data = ggstatsplot::movies_long,
genre %in% c("Action", "Action Comedy", "Action Drama", "Comedy")
),
cor.vars = length:votes,
colors = c("#cbac43", "white", "#550000"),
grouping.var = genre, # grouping variable
k = 3L, # number of digits after decimal point
title.prefix = "Movie genre",
messages = FALSE,
plotgrid.args = list(nrow = 2)
)
You can also get a dataframe containing all relevant details from the statistical tests:
# setup
set.seed(123)
# dataframe in long format
ggcorrmat(
data = ggplot2::msleep,
type = "bayes",
output = "dataframe"
)
#> # A tibble: 15 x 12
#> parameter1 parameter2 rho ci_low ci_high pd rope_percentage
#> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 sleep_total sleep_rem 0.735 0.617 0.810 1 0
#> 2 sleep_total sleep_cycle -0.436 -0.645 -0.194 0.998 0.0225
#> 3 sleep_total awake -1.00 -1.00 -1.00 1 0
#> 4 sleep_total brainwt -0.344 -0.525 -0.157 0.997 0.0222
#> 5 sleep_total bodywt -0.295 -0.456 -0.142 0.997 0.0318
#> 6 sleep_rem sleep_cycle -0.308 -0.539 -0.0463 0.969 0.0985
#> 7 sleep_rem awake -0.733 -0.827 -0.640 1 0
#> 8 sleep_rem brainwt -0.206 -0.413 0.0106 0.924 0.208
#> 9 sleep_rem bodywt -0.313 -0.492 -0.132 0.994 0.0368
#> 10 sleep_cycle awake 0.440 0.213 0.659 0.992 0.0205
#> 11 sleep_cycle brainwt 0.823 0.716 0.910 1 0
#> 12 sleep_cycle bodywt 0.379 0.133 0.607 0.988 0.0385
#> 13 awake brainwt 0.341 0.160 0.520 0.996 0.03
#> 14 awake bodywt 0.302 0.144 0.463 0.998 0.0295
#> 15 brainwt bodywt 0.925 0.892 0.955 1 0
#> prior_distribution prior_location prior_scale bf nobs
#> <chr> <dbl> <dbl> <dbl> <int>
#> 1 cauchy 0 0.707 3.00e+ 9 61
#> 2 cauchy 0 0.707 8.85e+ 0 32
#> 3 cauchy 0 0.707 NA 83
#> 4 cauchy 0 0.707 7.29e+ 0 56
#> 5 cauchy 0 0.707 9.28e+ 0 83
#> 6 cauchy 0 0.707 1.42e+ 0 32
#> 7 cauchy 0 0.707 3.01e+ 9 61
#> 8 cauchy 0 0.707 6.54e- 1 48
#> 9 cauchy 0 0.707 4.80e+ 0 61
#> 10 cauchy 0 0.707 8.85e+ 0 32
#> 11 cauchy 0 0.707 3.80e+ 6 30
#> 12 cauchy 0 0.707 3.76e+ 0 32
#> 13 cauchy 0 0.707 7.29e+ 0 56
#> 14 cauchy 0 0.707 9.27e+ 0 83
#> 15 cauchy 0 0.707 1.58e+22 56Following tests are carried out for each type of analyses. Additionally, the correlation coefficients (and their confidence intervals) are used as effect sizes-
| Type | Test | CI? |
|---|---|---|
| Parametric | Pearson’s correlation coefficient | Yes |
| Non-parametric | Spearman’s rank correlation coefficient | Yes |
| Robust | Percentage bend correlation coefficient | Yes |
| Bayes Factor | Pearson’s correlation coefficient | Yes |
For examples and more information, see the ggcorrmat vignette: https://indrajeetpatil.github.io/ggstatsplot/articles/web_only/ggcorrmat.html
ggcoefstatsThe function ggstatsplot::ggcoefstats generates dot-and-whisker plots for regression models saved in a tidy data frame. The tidy dataframes are prepared using the following packages: broom, broom.mixed, and parameters. Additionally, if available, the model summary indices are also extracted from the following packages: broom, broom.mixed, and performance.
Although the statistical models displayed in the plot may differ based on the class of models being investigated, there are few aspects of the plot that will be invariant across models:
The dot-whisker plot contains a dot representing the estimate and their confidence intervals (95% is the default). The estimate can either be effect sizes (for tests that depend on the F statistic) or regression coefficients (for tests with t and z statistic), etc. The function will, by default, display a helpful x-axis label that should clear up what estimates are being displayed. The confidence intervals can sometimes be asymmetric if bootstrapping was used.
The caption will always contain diagnostic information, if available, about models that can be useful for model selection: The smaller the Akaike’s Information Criterion (AIC) and the Bayesian Information Criterion (BIC) values, the “better” the model is.
The output of this function will be a ggplot2 object and, thus, it can be further modified (e.g., change themes, etc.) with ggplot2 functions.
# for reproducibility
set.seed(123)
# model
mod <- stats::lm(formula = mpg ~ am * cyl, data = mtcars)
# plot
ggstatsplot::ggcoefstats(mod)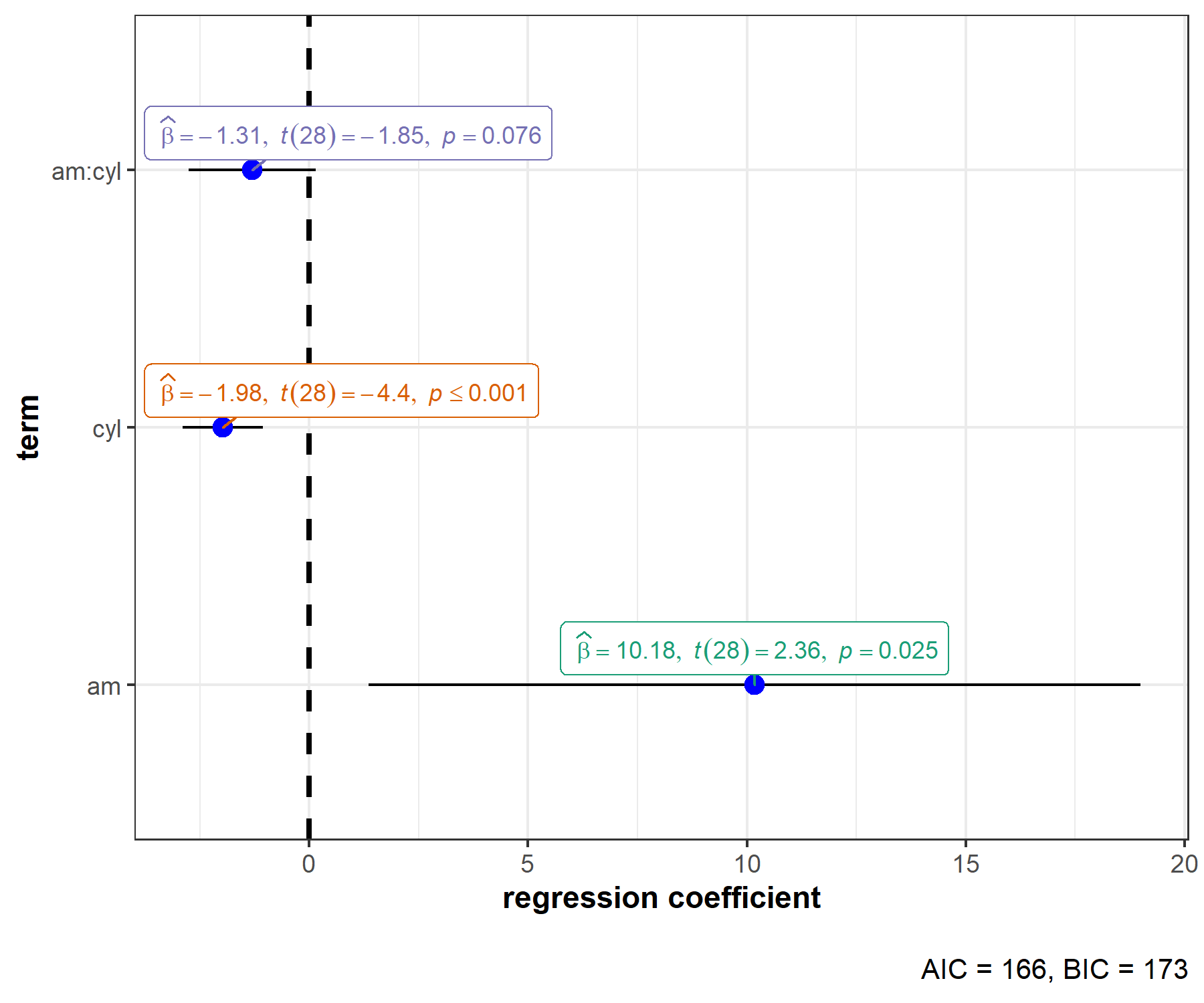
This default plot can be further modified to one’s liking with additional arguments (also, let’s use a different model now):
# for reproducibility
set.seed(123)
# plot
ggstatsplot::ggcoefstats(
x = MASS::rlm(formula = mpg ~ am * cyl, data = mtcars),
point.args = list(color = "red", size = 3, shape = 15),
vline.args = list(size = 1, color = "#CC79A7", linetype = "dotdash"),
stats.label.color = c("#0072B2", "#D55E00", "darkgreen"),
title = "Car performance predicted by transmission & cylinder count",
subtitle = "Source: 1974 Motor Trend US magazine",
ggtheme = hrbrthemes::theme_ipsum_ps(),
ggstatsplot.layer = FALSE
) + # note the order in which the labels are entered
ggplot2::scale_y_discrete(labels = c("transmission", "cylinders", "interaction")) +
ggplot2::labs(x = "regression coefficient", y = NULL)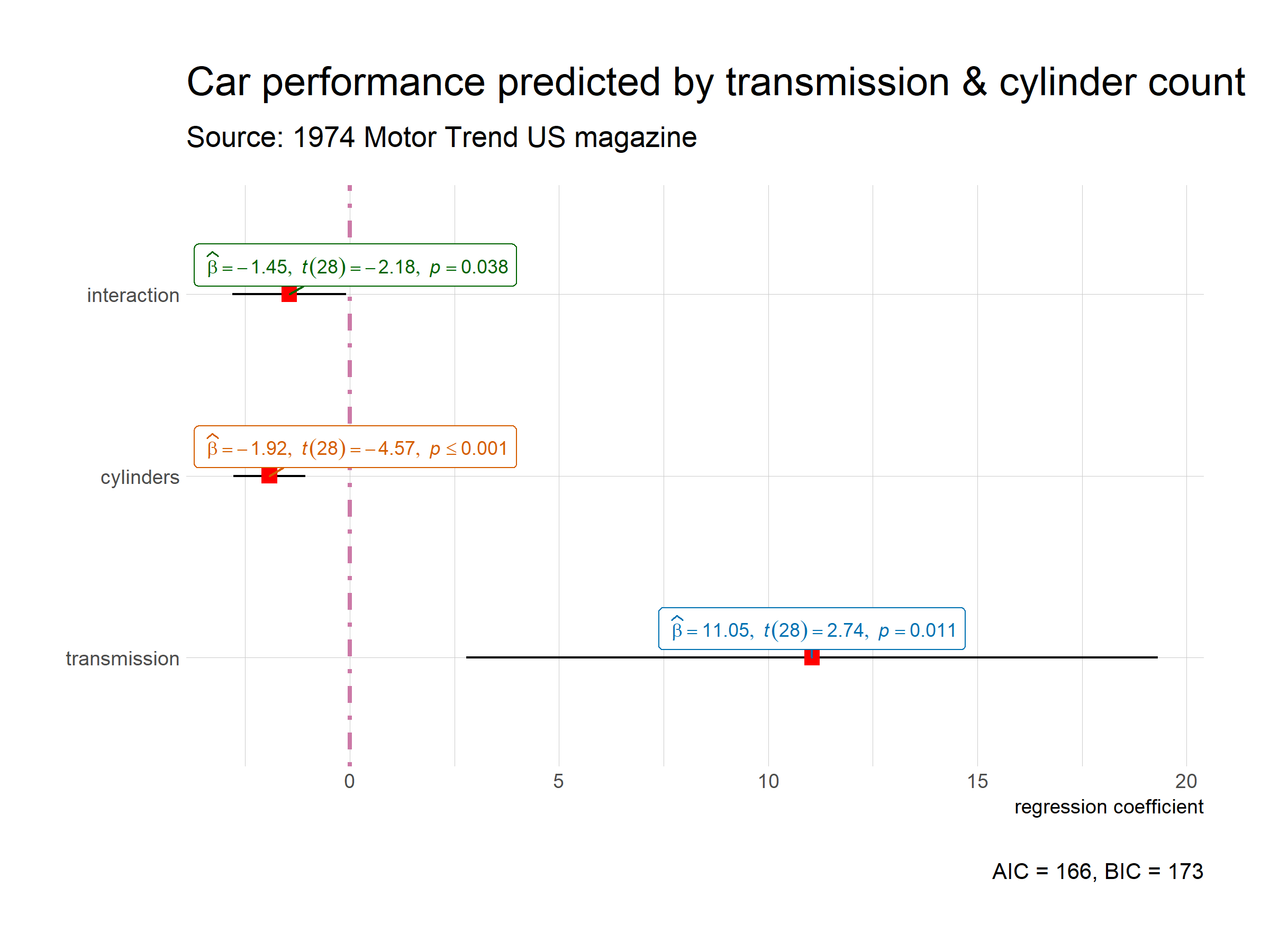
Most of the regression models that are supported in the underlying packages are also supported by ggcoefstats. For example-
aareg, anova, aov, aovlist, Arima, bayesx, bayesGARCH, BBmm, BBreg, bcplm, bglmerMod, bife, bigglm, biglm, blavaan, bmlm, blmerMod, bracl, brglm2, brmsfit, brmultinom, btergm, cch, cgam, cgamm, cglm, clm, clm2, clmm, clmm2, coeftest, complmrob, confusionMatrix, coxme, coxph, cpglm, cpglmm, crch, DirichReg, drc, emmGrid, epi.2by2, ergm, feis, felm, fitdistr, flexsurvreg, glmc, glmerMod, glmmTMB, gls, gam, Gam, gamlss, garch, glm, glmmadmb, glmmPQL, glmRob, glmrob, glmx, gmm, hurdle, ivreg, iv_robust, lavaan, lm, lm.beta, lmerMod, lmerModLmerTest, lmodel2, lmRob, lmrob, LORgee, lrm, mcmc, mcmc.list, MCMCglmm, mclogit, mmclogit, mediate, mixor, mjoint, mle2, mlm, multinom, negbin, nlmerMod, nlrq, nlreg, nls, orcutt, plm, polr, ridgelm, rjags, rlm, rlmerMod, robmixglm, rq, rqss, slm, speedglm, speedlm, stanfit, stanreg, survreg, svyglm, svyolr, svyglm, tobit, truncreg, vgam, wbgee, wblm, zcpglm, zeroinfl, etc.
Although not shown here, this function can also be used to carry out both frequentist and Bayesian random-effects meta-analysis.
For a more exhaustive account of this function, see the associated vignette- https://indrajeetpatil.github.io/ggstatsplot/articles/web_only/ggcoefstats.html
combine_plotsThe full power of ggstatsplot can be leveraged with a functional programming package like purrr that replaces for loops with code that is both more succinct and easier to read and, therefore, purrr should be preferrred 😻. (Another old school option to do this effectively is using the plyr package.)
In such cases, ggstatsplot contains a helper function combine_plots to combine multiple plots, which can be useful for combining a list of plots produced with purrr. This is a wrapper around cowplot::plot_grid and lets you combine multiple plots and add a combination of title, caption, and annotation texts with suitable defaults.
For examples (both with plyr and purrr), see the associated vignette- https://indrajeetpatil.github.io/ggstatsplot/articles/web_only/combine_plots.html
ggstatsplot statistical details with custom plotsSometimes you may not like the default plots produced by ggstatsplot. In such cases, you can use other custom plots (from ggplot2 or other plotting packages) and still use ggstatsplot functions to display results from relevant statistical test.
For example, in the following chunk, we will create plot (ridgeplot) using ggridges package and use ggstatsplot function for extracting results.
set.seed(123)
# loading the needed libraries
library(ggridges)
library(ggplot2)
library(ggstatsplot)
# using `ggstatsplot` to get call with statistical results
stats_results <-
ggstatsplot::ggbetweenstats(
data = morley,
x = Expt,
y = Speed,
output = "subtitle",
messages = FALSE
)
# using `ggridges` to create plot
ggplot(morley, aes(x = Speed, y = as.factor(Expt), fill = as.factor(Expt))) +
geom_density_ridges(
jittered_points = TRUE,
quantile_lines = TRUE,
scale = 0.9,
alpha = 0.7,
vline_size = 1,
vline_color = "red",
point_size = 0.4,
point_alpha = 1,
position = position_raincloud(adjust_vlines = TRUE)
) + # adding annotations
labs(
title = "Michelson-Morley experiments",
subtitle = stats_results,
x = "Speed of light",
y = "Experiment number"
) + # remove the legend
theme(legend.position = "none")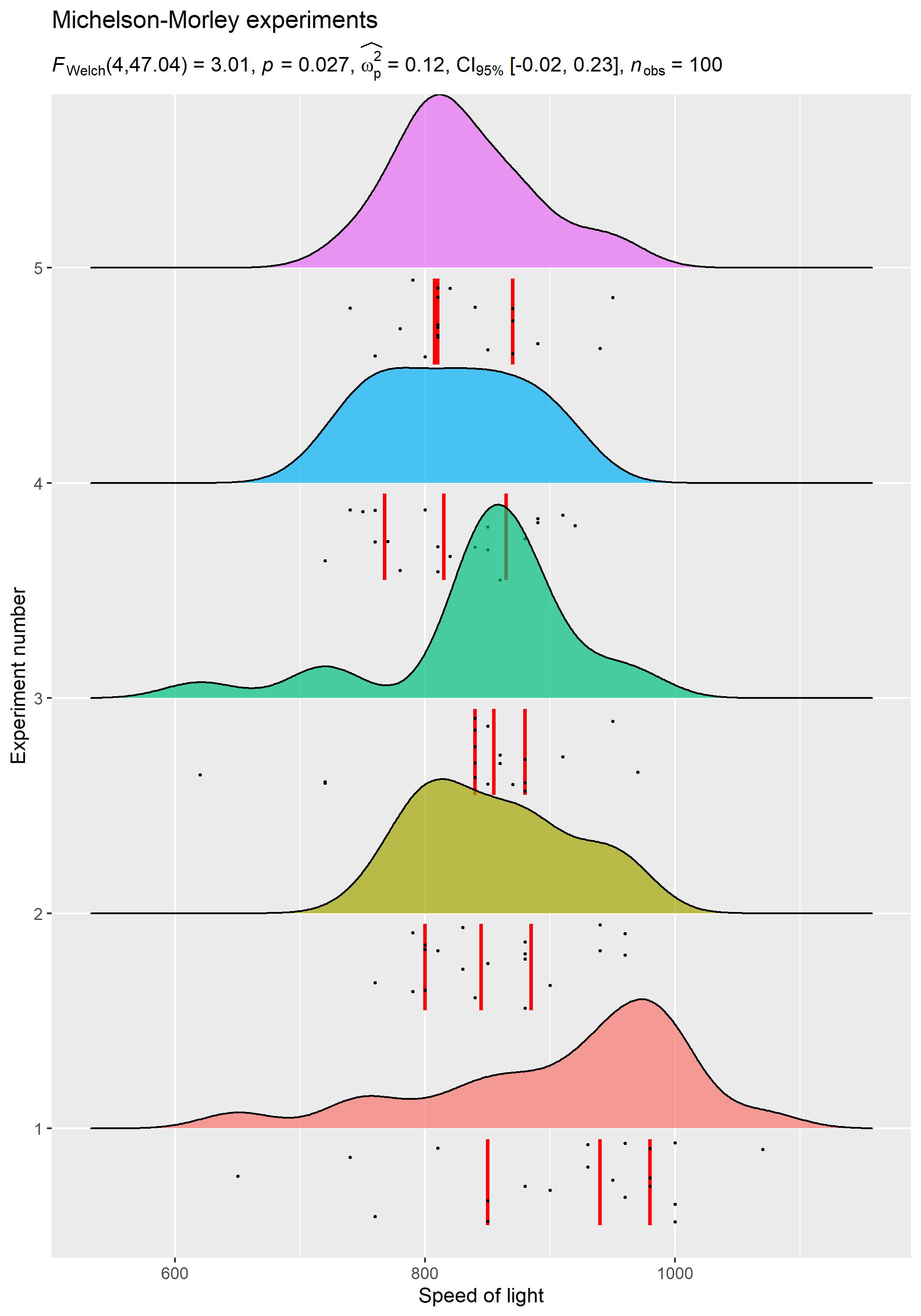
As seen from these examples, ggstatsplot relies on non-standard evaluation (NSE) - implemented via rlang - i.e., rather than looking at the values of arguments (x, y), it instead looks at their expressions. Therefore, the syntax is simpler and follows the following principles-
data argument must always be specified.$ operator cannot be used to specify variables in a dataframe.x = "var1") and symbol (x = var1) arguments.These set principles combined with the fact that almost all functions produce publication-ready plots that require very few arguments if one finds the aesthetic and statistical defaults satisfying make the syntax much less cognitively demanding and easy to remember/reconstruct.
ggstatsplot is a very chatty package and will by default print helpful notes on assumptions about statistical tests, warnings, etc. If you don’t want your console to be cluttered with such messages, they can be turned off by setting argument messages = FALSE in the function call.
Most functions share a type (of test) argument that is helpful to specify the type of statistical analysis:
"p" (for parametric)"np" (for non-parametric)"r" (for robust)"bf" (for Bayes Factor)All relevant functions in ggstatsplot have a output argument which can be used to not only return plots (which is the default), but also to return a subtitle or caption, which are objects of type call and can be used to display statistical details in conjunction with a custom plot and at a custom location in the plot.
Additionally, all functions share the ggtheme and palette arguments that can be used to specify your favorite ggplot theme and color palette.
ggstatsverse: Components of ggstatsplotTo make the maintenance and development of ggstatsplot more manageable, it is being broken into smaller pieces. Currently, the package internally relies on the following packages that manage different aspects of statistical analyses:

statsExpressionsThe statsExpressions package forms the statistical backend that processes data and creates expressions containing results from statistical tests and are by default displayed in as plot subtitle.
For more exhaustive documentation for this package, see: https://indrajeetpatil.github.io/statsExpressions/
tidyBFThe tidyBF package forms the backend that processes data and creates expressions containing results from Bayes Factor tests and are by default displayed in as plot caption.
For more exhaustive documentation for this package, see: https://indrajeetpatil.github.io/tidyBF/
pairwiseComparisonsThe pairwiseComparisons package forms the pairwise comparison backend for creating results that are used to display post hoc multiple comparisons displayed in ggbetweenstats and ggwithinstats functions.
For more exhaustive documentation for this package, see: https://indrajeetpatil.github.io/pairwiseComparisons/
ipmiscThe ipmisc package contains some of the data wrangling/cleaning functions and a few other miscellaneous functions.
For more exhaustive documentation for this package, see: https://indrajeetpatil.github.io/ipmisc/
I would like to thank all the contributors to ggstatsplot who pointed out bugs or requested features I hadn’t considered. I would especially like to thank Chuck Powell for his initial contributions to the package. I also want to thank other package maintainers (like Daniel Lüdecke, Dominique Makowski, Patrick Mair, Salvatore Mangiafico, etc.) who have patiently and diligently answered my relentless number of questions and added feature requests I wanted.
The hexsticker was generously designed by Sarah Otterstetter (Max Planck Institute for Human Development, Berlin). This package has also benefited from the larger rstats community on Twitter and StackOverflow.
Thanks are also due to my postdoc advisers (Mina Cikara and Fiery Cushman at Harvard; Iyad Rahwan at Max Planck Institute for Human Development) who patiently supported me spending hundreds of hours working on this package rather than what I was paid to do. 😄
As the code stands right now, here is the code coverage for all primary functions involved: https://codecov.io/gh/IndrajeetPatil/ggstatsplot/tree/master/R
I’m happy to receive bug reports, suggestions, questions, and (most of all) contributions to fix problems and add features. I personally prefer using the GitHub issues system over trying to reach out to me in other ways (personal e-mail, Twitter, etc.). Pull Requests for contributions are encouraged.
Here are some simple ways in which you can contribute (in the increasing order of commitment):
Read and correct any inconsistencies in the documentation
Raise issues about bugs or wanted features
Review code
Add new functionality (in the form of new plotting functions or helpers for preparing subtitles)
Please note that this project is released with a Contributor Code of Conduct. By participating in this project you agree to abide by its terms.
For reproducibility purposes, the details about the session information in which this document was rendered, see- https://indrajeetpatil.github.io/ggstatsplot/articles/web_only/session_info.html