europepmc facilitates access to the Europe PMC RESTful Web Service. The client furthermore supports the Europe PMC Annotations API to retrieve text-mined concepts and terms per article.
Europe PMC covers life science literature and gives access to open access full texts. Europe PMC ingests all PubMed content and extends its index with other literature and patent sources.
For more infos on Europe PMC, see:
Levchenko, M., Gou, Y., Graef, F., Hamelers, A., Huang, Z., Ide-Smith, M., … McEntyre, J. (2017). Europe PMC in 2017. Nucleic Acids Research, 46(D1), D1254–D1260. https://doi.org/10.1093/nar/gkx1005
This client supports the following API methods from the Articles RESTful API:
| API-Method | Description | R functions |
|---|---|---|
| search | Search Europe PMC and get detailed metadata | epmc_search(), epmc_details(), epmc_search_by_doi() |
| profile | Obtain a summary of hit counts for several Europe PMC databases | epmc_profile() |
| citations | Load metadata representing citing articles for a given publication | epmc_citations() |
| references | Retrieve the reference section of a publication | epmc_refs() |
| databaseLinks | Get links to biological databases such as UniProt or ENA | epmc_db(), epmc_db_count() |
| labslinks | Access links to Europe PMC provided by third parties | epmc_lablinks(), epmc_lablinks_count() |
| fullTextXML | Fetch full-texts deposited in PMC | epmc_ftxt() |
| bookXML | retrieve book XML formatted full text for the Open Access subset of the Europe PMC bookshelf | epmc_ftxt_book() |
From the Europe PMC Annotations API:
| API-Method | Description | R functions |
|---|---|---|
| annotationsByArticleIds | Get the annotations contained in the list of articles specified | epmc_annotations_by_id() |
From CRAN
The latest development version can be installed using the remotes package:
Loading into R
The search covers both metadata (e.g. abstracts or title) and full texts. To build your query, please refer to the comprehensive guidance on how to search Europe PMC: http://europepmc.org/help. Provide your query in the Europe PMC search syntax to epmc_search().
europepmc::epmc_search(query = '"2019-nCoV" OR "2019nCoV"')
#> # A tibble: 100 x 29
#> id source pmid pmcid doi title authorString journalTitle pubYear journalIssn pubType isOpenAccess
#> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
#> 1 3240… MED 3240… PMC7… 10.1… Livi… Santillan-G… Med Intensi… 2020 "0210-5691… letter Y
#> 2 PPR1… PPR <NA> <NA> 10.1… The … Benvenuto D… <NA> 2020 <NA> prepri… N
#> 3 3203… MED 3203… PMC7… 10.1… Emer… Malik YS, S… Vet Q 2020 "0165-2176… other;… Y
#> 4 3238… MED 3238… PMC7… 10.1… Comp… Yu R, Chen … Int J Antim… 2020 "1872-7913… resear… Y
#> 5 3230… MED 3230… PMC7… 10.1… A ca… Yang X, Zha… Clin Res He… 2020 "2210-7401… case r… Y
#> 6 3203… MED 3203… PMC7… 10.1… Coro… Bonilla-Ald… Travel Med … 2020 "1477-8939… resear… Y
#> 7 3220… MED 3220… PMC7… 10.4… Ther… Sarma P, Pr… Indian J Ph… 2020 "0253-7613… editor… Y
#> 8 3217… MED 3217… PMC7… 10.1… Anes… Zhao S, Lin… J Cardiotho… 2020 "1053-0770… resear… Y
#> 9 3240… MED 3240… PMC7… 10.1… Prev… Wang X, Wan… Diabetes Re… 2020 "0168-8227… resear… Y
#> 10 PMC7… PMC <NA> PMC7… 10.1… The … Rastogi YR,… Int J Envir… <NA> "1735-1472… review… Y
#> # … with 90 more rows, and 17 more variables: inEPMC <chr>, inPMC <chr>, hasPDF <chr>, hasBook <chr>,
#> # hasSuppl <chr>, citedByCount <int>, hasReferences <chr>, hasTextMinedTerms <chr>,
#> # hasDbCrossReferences <chr>, hasLabsLinks <chr>, hasTMAccessionNumbers <chr>, firstIndexDate <chr>,
#> # firstPublicationDate <chr>, issue <chr>, journalVolume <chr>, pageInfo <chr>, versionNumber <int>Be aware that Europe PMC expands queries with MeSH synonyms by default. You can turn this behavior off using the synonym = FALSE parameter.
By default, epmc_search() returns 100 records. To adjust the limit, simply use the limit parameter.
See vignette Introducing europepmc, an R interface to Europe PMC RESTful API for a long-form documentation about how to search Europe PMC with this client.
epmc_hits_trend()There is also a nice function allowing you to easily create review graphs like described in Maëlle Salmon’s blog post:
tt_oa <- europepmc::epmc_hits_trend("Malaria", period = 1995:2019, synonym = FALSE)
tt_oa
#> # A tibble: 25 x 3
#> year all_hits query_hits
#> <int> <dbl> <dbl>
#> 1 1995 448961 1486
#> 2 1996 458444 1560
#> 3 1997 456594 1857
#> 4 1998 474525 1748
#> 5 1999 493574 1935
#> 6 2000 531892 2130
#> 7 2001 545533 2204
#> 8 2002 561118 2355
#> 9 2003 588172 2588
#> 10 2004 627729 2806
#> # … with 15 more rows
# we use ggplot2 for plotting the graph
library(ggplot2)
ggplot(tt_oa, aes(year, query_hits / all_hits)) +
geom_point() +
geom_line() +
xlab("Year published") +
ylab("Proportion of articles on Malaria in Europe PMC")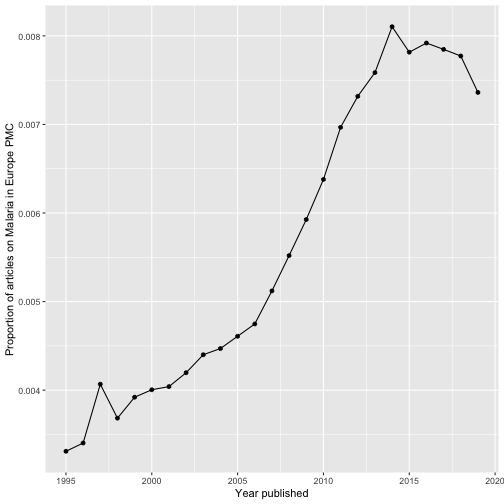
For more info, read the vignette about creating literature review graphs:
https://docs.ropensci.org/europepmc/articles/evergreenreviewgraphs.html
Check out the tidypmc package
https://github.com/ropensci/tidypmc
The package maintainer, Chris Stubben (@cstubben), has also created an Shiny App that allows you to search and browse Europe PMC:
https://cstubben.shinyapps.io/euPMC/
oai to get metadata and full text via Europe PMC’s OAI interface: https://github.com/ropensci/oairentrez to interact with NCBI databases such as PubMed: https://github.com/ropensci/rentrezfulltext package gives access to supplementary material of open access life-science publications in Europe PMC: https://github.com/ropensci/fulltextPlease note that this project is released with a Contributor Code of Conduct. By participating in this project you agree to abide by its terms.
License: GPL-3
Please use the issue tracker for bug reporting and feature requests.