ℹ️ Tutorials ℹ️ Reference documentation

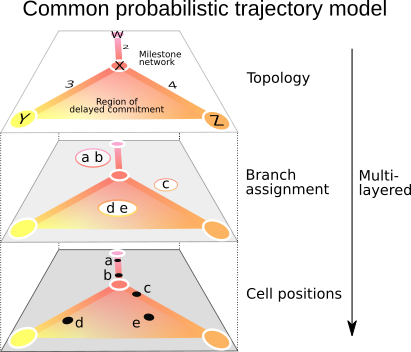
dynwrap contains the code for a common model of single-cell trajectories. The package can:
Documentation and the API reference for dynwrap can be found at the dyvnerse documentation website: https://dynverse.org/ .
dynwrap was used to wrap 50+ trajectory inference method within docker containers in dynmethods.
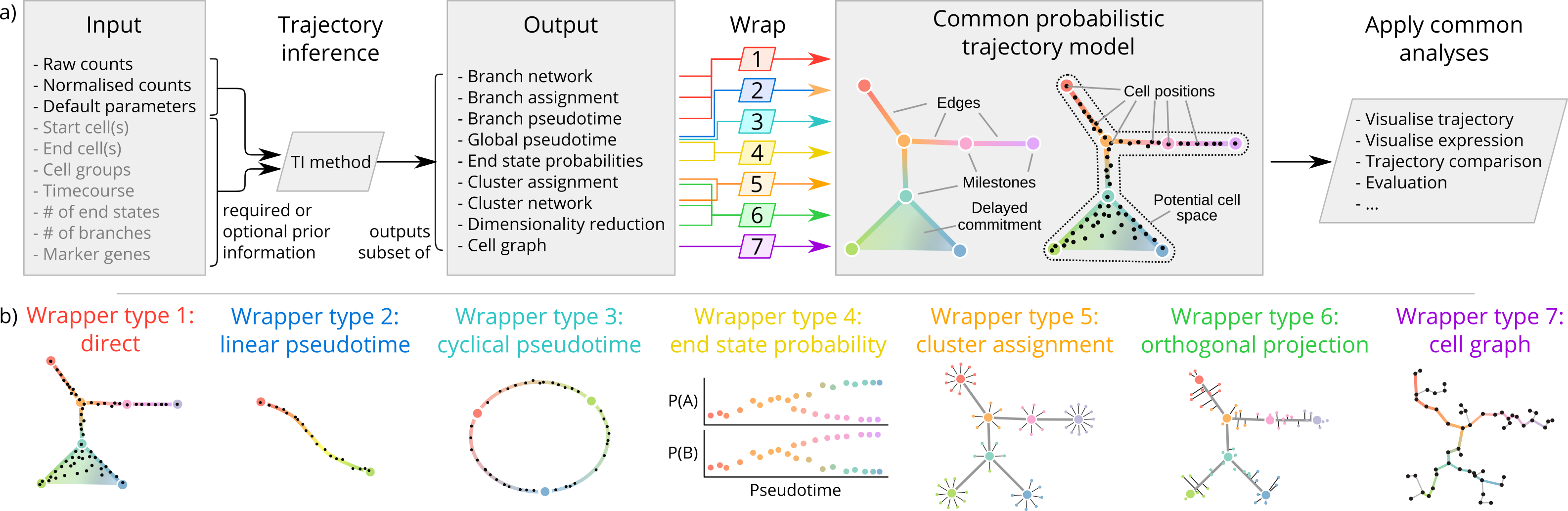
The advantage of using a common model is that it allows:
Check out news(package = "dynwrap") or NEWS.md for a full list of changes.
BUG FIX project_trajectory(): Correctly pass parameters.
MINOR CHANGES select_waypoints(): Do not recompute waypoints if trajectory already contains some.
MINOR CHANGES convert_progressions_to_milestone_percentages(): Solve tapply issues ahead of dplyr 1.0 release.
FUNCTIONALITY: Improved RNA velocity handling. Not all features need to be present in the projected expression, allowing integration with standard velocyto.R pipelines.
BUG FIX calculate_pseudotime(): Fix pseudotime calculation on branching trajectories (#139).
BUG FIX flip_edges(): Also flip dimred_segment_progressions if available.
BUG FIX add_root(): Fix rooting for some linear trajectories (#149)
MINOR CHANGES: Allow a named vector for groups_id prior information (#154)
MINOR CHANGES: Moved orient_topology_to_velocity() to scvelo R package.
BUG FIX simplify_trajectory(): handle dimred_milestones and dimred_segments values (#153).
BUG FIX add_dimred(): Do not execute example of dyndimred if is not installed.