Density ratio estimation is described as follows: for given two data samples x1 and x2 from unknown distributions p(x) and q(x) respectively, estimate w(x) = p(x) / q(x), where x1 and x2 are d-dimensional real numbers.
The estimated density ratio function w(x) can be used in many applications such as anomaly detection [Hido et al. 2011], change-point detection [Liu et al. 2013], and covariate shift adaptation [Sugiyama et al. 2007]. Other useful applications about density ratio estimation were summarized by [Sugiyama et al. 2012].
The package densratio provides a function densratio() that returns an object with a method to estimate density ratio as compute_density_ratio().
For example,
set.seed(3)
x1 <- rnorm(200, mean = 1, sd = 1/8)
x2 <- rnorm(200, mean = 1, sd = 1/2)
library(densratio)
densratio_obj <- densratio(x1, x2)The densratio object has a function compute_density_ratio() that can compute the estimated density ratio w_hat(x) for any d-dimensional input x (now d=1).
new_x <- seq(0, 2, by = 0.03)
w_hat <- densratio_obj$compute_density_ratio(new_x)
plot(new_x, w_hat, pch=19)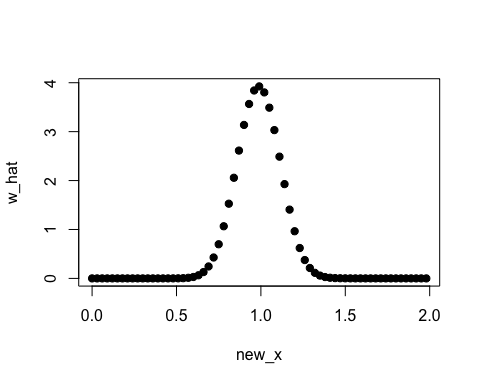
In this case, the true density ratio w(x) = p(x)/q(y) = Norm(1, 1/8) / Norm(1, 1/2) is known. So we can compare w(x) with the estimated density ratio w-hat(x).
true_density_ratio <- function(x) dnorm(x, 1, 1/8) / dnorm(x, 1, 1/2)
plot(true_density_ratio, xlim=c(0, 2), lwd=2, col="red", xlab = "x", ylab = "Density Ratio")
plot(densratio_obj$compute_density_ratio, xlim=c(0, 2), lwd=2, col="green", add=TRUE)
legend("topright", legend=c(expression(w(x)), expression(hat(w)(x))), col=2:3, lty=1, lwd=2, pch=NA)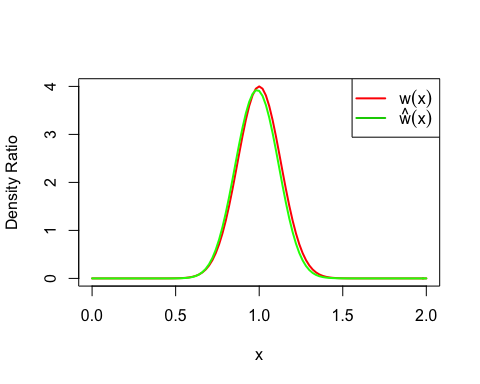
You can install the densratio package from CRAN.
You can also install the package from GitHub.
install.packages("remotes") # if you have not installed "remotes" package
remotes::install_github("hoxo-m/densratio")The source code for densratio package is available on GitHub at
The package provides densratio(). The function returns an object that has a function to compute estimated density ratio.
For data samples x1 and x2,
set.seed(3)
x1 <- rnorm(200, mean = 1, sd = 1/8)
x2 <- rnorm(200, mean = 1, sd = 1/2)
library(densratio)
densratio_obj <- densratio(x1, x2)In this case, densratio_obj$compute_density_ratio() can compute estimated density ratio.
new_x <- seq(0, 2, by = 0.03)
w_hat <- densratio_obj$compute_density_ratio(new_x)
plot(new_x, w_hat, pch=19)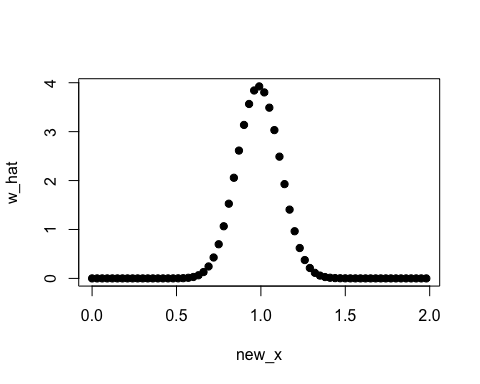
densratio() has method argument that you can pass "uLSIF", "RuSLIF", or "KLIEP".
The methods assume that density ratio are represented by linear model:
w(x) = theta_1 * K(x, c_1) + theta_2 * K(x, c_2) + ... + theta_b * K(x, c_b)where
K(x, c) = exp(-||x - c||^2 / 2 * sigma^2)is the Gaussian (RBF) kernel.
densratio() performs the following:
sigma by cross-validation,theta (in other words, find the optimal coefficients of the linear model), andsigma and theta are saved into densratio object, and are used when to compute density ratio in the call compute_density_ratio().densratio() outputs the result like as follows:
#>
#> Call:
#> densratio(x = x1, y = x2, method = "uLSIF")
#>
#> Kernel Information:
#> Kernel type: Gaussian
#> Number of kernels: 100
#> Bandwidth (sigma): 0.1
#> Centers: num [1:100, 1] 0.907 1.093 1.18 1.136 1.046 ...
#>
#> Kernel Weights:
#> num [1:100] 0.067455 0.040045 0.000459 0.016849 0.067084 ...
#>
#> Regularization Parameter (lambda): 1
#>
#> Function to Estimate Density Ratio:
#> compute_density_ratio()kernel_num argument. In default, kernel_num = 100.sigma = "auto", the algorithm automatically select an optimal value by cross validation. If you set sigma a number, that will be used. If you set sigma a numeric vector, the algorithm select an optimal value in them by cross validation.x1 underlying a numerator distribution p(x). You can find the whole values in result$kernel_info$centers.theta parameters in the linear kernel model. You can find these values in result$kernel_weights.compute_density_ratio().So far, the input data samples x1 and x2 were one dimensional. densratio() allows to input multidimensional data samples as matrix, as long as their dimensions are the same.
For example,
library(densratio)
library(mvtnorm)
set.seed(3)
x1 <- rmvnorm(300, mean = c(1, 1), sigma = diag(1/8, 2))
x2 <- rmvnorm(300, mean = c(1, 1), sigma = diag(1/2, 2))
densratio_obj_2d <- densratio(x1, x2)
densratio_obj_2d
#>
#> Call:
#> densratio(x = x1, y = x2, method = "uLSIF")
#>
#> Kernel Information:
#> Kernel type: Gaussian
#> Number of kernels: 100
#> Bandwidth(sigma): 0.316
#> Centers: num [1:100, 1:2] 1.257 0.758 1.122 1.3 1.386 ...
#>
#> Kernel Weights:
#> num [1:100] 0.0756 0.0986 0.059 0.0797 0.0421 ...
#>
#> Regularization Parameter (lambda): 0.3162278
#>
#> Function to Estimate Density Ratio:
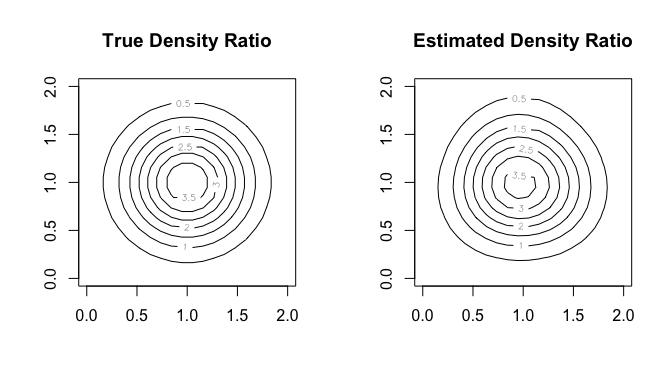
#> compute_density_ratio()In this case, as well, we can compare the true density ratio with the estimated density ratio.
true_density_ratio <- function(x) {
dmvnorm(x, mean = c(1, 1), sigma = diag(1/8, 2)) /
dmvnorm(x, mean = c(1, 1), sigma = diag(1/2, 2))
}
N <- 20
range <- seq(0, 2, length.out = N)
input <- expand.grid(range, range)
w_true <- matrix(true_density_ratio(input), nrow = N)
w_hat <- matrix(densratio_obj_2d$compute_density_ratio(input), nrow = N)
par(mfrow = c(1, 2))
contour(range, range, w_true, main = "True Density Ratio")
contour(range, range, w_hat, main = "Estimated Density Ratio")