

The coronavirus package provides a tidy format dataset of the 2019 Novel Coronavirus COVID-19 (2019-nCoV) epidemic. The raw data pulled from the Johns Hopkins University Center for Systems Science and Engineering (JHU CCSE) Coronavirus repository.
More details available here, and a csv format of the package dataset available here
A summary dashboard is available here
Source: Centers for Disease Control and Prevention’s Public Health Image Library
As this an ongoing situation, frequent changes in the data format may occur, please visit the package news to get updates about those changes
Install the CRAN version:
Install the Github version (refreshed on a daily bases):
While the coronavirus CRAN version is updated every month or two, the Github (Dev) version is updated on a daily bases. The update_dataset function enables to overcome this gap and keep the installed version with the most recent data available on the Github version:
Note: must restart the R session to have the updates available
Alternatively, you can pull the data using the Covid19R project data standard format with the refresh_coronavirus_jhu function:
covid19_df <- refresh_coronavirus_jhu()
head(covid19_df)
#> date location location_type location_code location_code_type data_type value lat long
#> 1 2020-05-30 Afghanistan country AF iso_3166_2 deaths_new 3 33.93911 67.709953
#> 2 2020-05-27 Afghanistan country AF iso_3166_2 deaths_new 7 33.93911 67.709953
#> 3 2020-05-29 Afghanistan country AF iso_3166_2 deaths_new 11 33.93911 67.709953
#> 4 2020-02-01 Afghanistan country AF iso_3166_2 cases_new 0 33.93911 67.709953
#> 5 2020-05-26 Afghanistan country AF iso_3166_2 deaths_new 1 33.93911 67.709953
#> 6 2020-06-01 Afghanistan country AF iso_3166_2 deaths_new 8 33.93911 67.709953This coronavirus dataset has the following fields:
date - The date of the summaryprovince - The province or state, when applicablecountry - The country or region namelat - Latitude pointlong - Longitude pointtype - the type of case (i.e., confirmed, death)cases - the number of daily cases (corresponding to the case type)head(coronavirus)
#> date province country lat long type cases
#> 1 2020-01-22 Afghanistan 33.93911 67.709953 confirmed 0
#> 2 2020-01-23 Afghanistan 33.93911 67.709953 confirmed 0
#> 3 2020-01-24 Afghanistan 33.93911 67.709953 confirmed 0
#> 4 2020-01-25 Afghanistan 33.93911 67.709953 confirmed 0
#> 5 2020-01-26 Afghanistan 33.93911 67.709953 confirmed 0
#> 6 2020-01-27 Afghanistan 33.93911 67.709953 confirmed 0Summary of the total confrimed cases by country (top 20):
library(dplyr)
summary_df <- coronavirus %>%
filter(type == "confirmed") %>%
group_by(country) %>%
summarise(total_cases = sum(cases)) %>%
arrange(-total_cases)
summary_df %>% head(20)
#> # A tibble: 20 x 2
#> country total_cases
#> <chr> <int>
#> 1 US 3711413
#> 2 Brazil 2074860
#> 3 India 1077781
#> 4 Russia 764215
#> 5 South Africa 350879
#> 6 Peru 349500
#> 7 Mexico 338913
#> 8 Chile 328846
#> 9 United Kingdom 295632
#> 10 Iran 271606
#> 11 Pakistan 263496
#> 12 Spain 260255
#> 13 Saudi Arabia 248416
#> 14 Italy 244216
#> 15 Turkey 218717
#> 16 France 211943
#> 17 Germany 202426
#> 18 Bangladesh 202066
#> 19 Colombia 190700
#> 20 Argentina 122524Summary of new cases during the past 24 hours by country and type (as of 2020-07-18):
library(tidyr)
coronavirus %>%
filter(date == max(date)) %>%
select(country, type, cases) %>%
group_by(country, type) %>%
summarise(total_cases = sum(cases)) %>%
pivot_wider(names_from = type,
values_from = total_cases) %>%
arrange(-confirmed)
#> # A tibble: 188 x 4
#> # Groups: country [188]
#> country confirmed death recovered
#> <chr> <int> <int> <int>
#> 1 US 63698 853 15516
#> 2 India 38697 543 23672
#> 3 Brazil 28532 921 18888
#> 4 South Africa 13285 144 4047
#> 5 Kyrgyzstan 11505 727 6883
#> 6 Colombia 8560 228 5199
#> 7 Mexico 7615 578 7037
#> 8 Russia 6214 122 7442
#> 9 Peru 3963 199 4104
#> 10 Argentina 3223 42 2827
#> 11 Bangladesh 2709 34 1373
#> 12 Saudi Arabia 2565 40 3057
#> 13 Chile 2407 98 2635
#> 14 Philippines 2303 113 319
#> 15 Iran 2166 188 2427
#> 16 Iraq 2049 75 1997
#> 17 Bolivia 2036 57 318
#> 18 Israel 1906 9 604
#> 19 Indonesia 1752 59 1434
#> 20 Pakistan 1579 46 5767
#> 21 Dominican Republic 1406 29 184
#> 22 Oman 1311 10 1322
#> 23 Honduras 1048 34 96
#> 24 Ecuador 938 32 353
#> 25 Turkey 918 17 1179
#> 26 Romania 889 21 176
#> 27 Ukraine 867 23 804
#> 28 Panama 853 33 974
#> 29 Japan 842 1 267
#> 30 United Kingdom 829 40 10
#> 31 Egypt 698 63 566
#> 32 Kenya 688 3 457
#> 33 Kuwait 683 3 639
#> 34 Nigeria 653 6 305
#> 35 Algeria 601 11 314
#> 36 Costa Rica 582 7 84
#> 37 Uzbekistan 579 4 124
#> 38 Bahrain 531 0 577
#> 39 Azerbaijan 497 8 645
#> 40 Ghana 488 1 129
#> # … with 148 more rowsPlotting the total cases by type worldwide:
library(plotly)
coronavirus %>%
group_by(type, date) %>%
summarise(total_cases = sum(cases)) %>%
pivot_wider(names_from = type, values_from = total_cases) %>%
arrange(date) %>%
mutate(active = confirmed - death - recovered) %>%
mutate(active_total = cumsum(active),
recovered_total = cumsum(recovered),
death_total = cumsum(death)) %>%
plot_ly(x = ~ date,
y = ~ active_total,
name = 'Active',
fillcolor = '#1f77b4',
type = 'scatter',
mode = 'none',
stackgroup = 'one') %>%
add_trace(y = ~ death_total,
name = "Death",
fillcolor = '#E41317') %>%
add_trace(y = ~recovered_total,
name = 'Recovered',
fillcolor = 'forestgreen') %>%
layout(title = "Distribution of Covid19 Cases Worldwide",
legend = list(x = 0.1, y = 0.9),
yaxis = list(title = "Number of Cases"),
xaxis = list(title = "Source: Johns Hopkins University Center for Systems Science and Engineering"))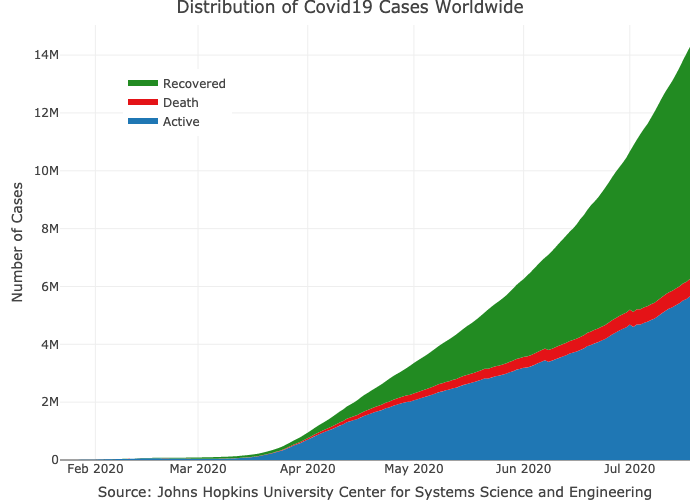
Plot the confirmed cases distribution by counrty with treemap plot:
conf_df <- coronavirus %>%
filter(type == "confirmed") %>%
group_by(country) %>%
summarise(total_cases = sum(cases)) %>%
arrange(-total_cases) %>%
mutate(parents = "Confirmed") %>%
ungroup()
plot_ly(data = conf_df,
type= "treemap",
values = ~total_cases,
labels= ~ country,
parents= ~parents,
domain = list(column=0),
name = "Confirmed",
textinfo="label+value+percent parent")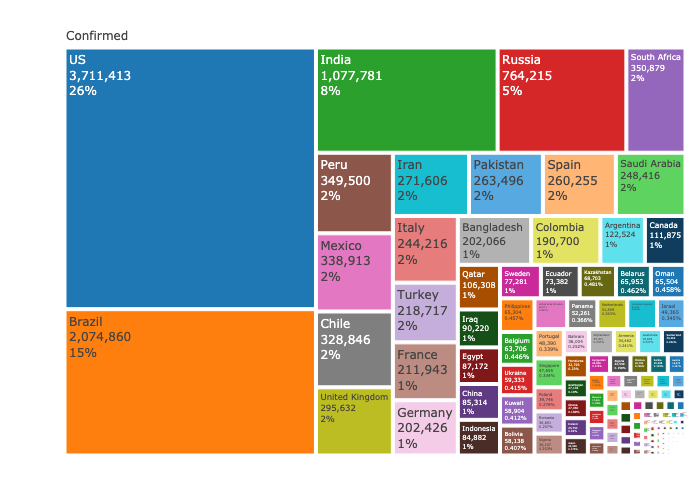
The raw data pulled and arranged by the Johns Hopkins University Center for Systems Science and Engineering (JHU CCSE) from the following resources: