dplyrpins
DBI connectionsThe main goal of connections is to integrate DBI-compliant packages with the RStudio IDE’s Connection Pane. Packages such as RPostgres, RSQLite, RMariaDB and bigrquery connect R to those databases, but do not provide a direct integration with the Connections Pane. connections reads the configuration of the connection and creates the integration with RStudio.
A second goal is to provide integration with the pins package. The connections package allows you to pin database connections and dplyr table objects.
Install the development version from GitHub with:
The two main functions added by connections are:
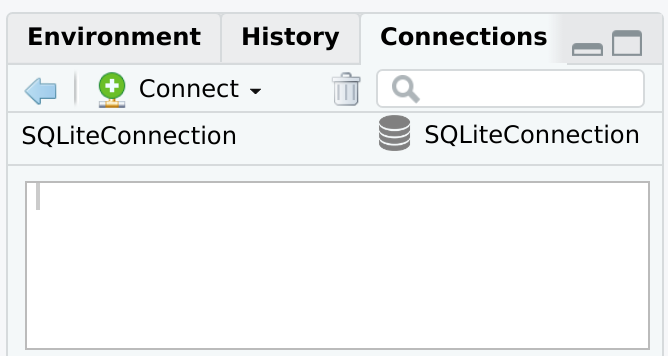
connection_open() - Opens the database connection. Use instead of dbConnect(), but use the exact same arguments. It also automatically starts the Connections pane.connection_close() - Closes the database connection.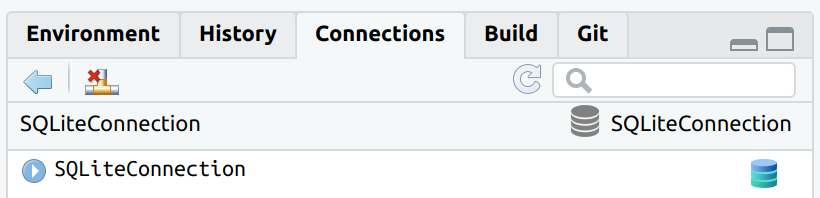
The connection can now be closed by using the appropriate button in the Connections pane, or by using connection_close()
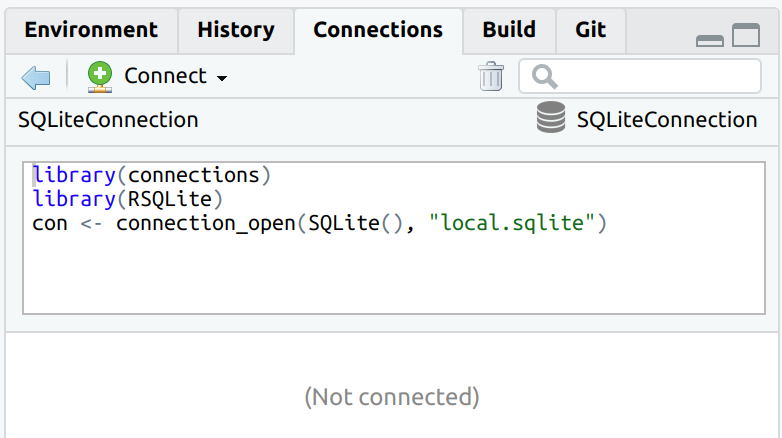
The connection code is parsed when connecting to the database, and it is visible once the connection is closed.
dplyrconnections integrates with dplyr by supporting the following two functions:
tbl() - To create a pointer to a table or view within the database.copy_to() - To copy data from the R session to the database.The version of copy_to() inside connections automatically updates the Connections pane, so the new table automatically shows up.
con <- connection_open(SQLite(), "local.sqlite")
copy_to(con, mtcars, temporary = FALSE, overwrite = TRUE)
#> # Source: table<mtcars> [?? x 11]
#> # Database: sqlite 3.29.0 [/home/edgar/connections/local.sqlite]
#> mpg cyl disp hp drat wt qsec vs am gear carb
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 21 6 160 110 3.9 2.62 16.5 0 1 4 4
#> 2 21 6 160 110 3.9 2.88 17.0 0 1 4 4
#> 3 22.8 4 108 93 3.85 2.32 18.6 1 1 4 1
#> 4 21.4 6 258 110 3.08 3.22 19.4 1 0 3 1
#> 5 18.7 8 360 175 3.15 3.44 17.0 0 0 3 2
#> 6 18.1 6 225 105 2.76 3.46 20.2 1 0 3 1
#> 7 14.3 8 360 245 3.21 3.57 15.8 0 0 3 4
#> 8 24.4 4 147. 62 3.69 3.19 20 1 0 4 2
#> 9 22.8 4 141. 95 3.92 3.15 22.9 1 0 4 2
#> 10 19.2 6 168. 123 3.92 3.44 18.3 1 0 4 4
#> # … with more rowsTo use an existing table inside the database use tbl().
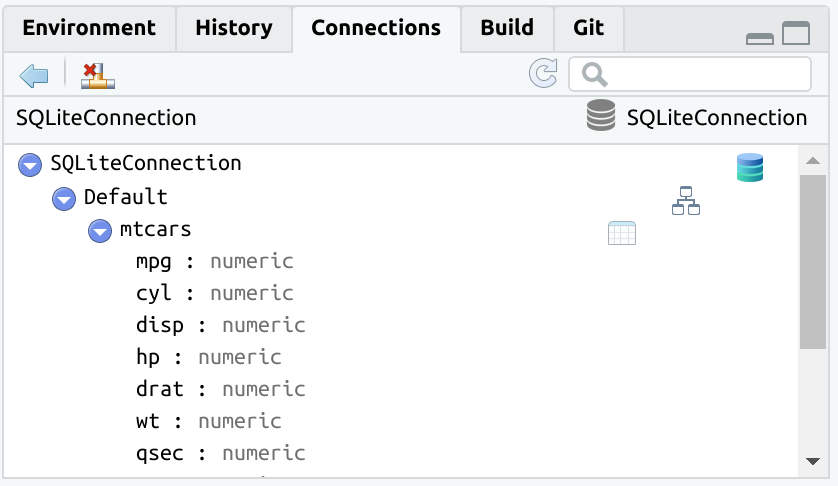
The tbl() function opens the rest of the already available dplyr database integration.
db_mtcars %>%
group_by(am) %>%
summarise(avg_mpg = mean(mpg, na.rm = TRUE))
#> # Source: lazy query [?? x 2]
#> # Database: sqlite 3.29.0 [/home/edgar/connections/local.sqlite]
#> am avg_mpg
#> <dbl> <dbl>
#> 1 0 17.1
#> 2 1 24.4pinsThe connections package integrates with pins. It enables the ability to save and retrieve connections and queries.
Use the same pin() command to save a database connection. Under the hood, connections saves the necessary information to recreate the connection code, not the actual connection R object.
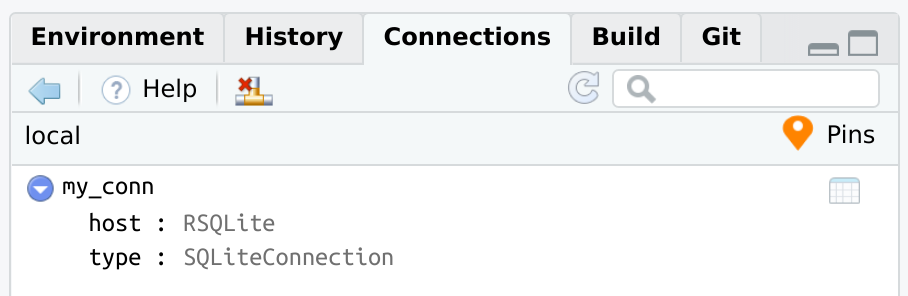
Use pin_get() to re-open the connection. In effect, pin_get() will replay the exact same code used to initially connect to the database. This means that connection_open() is already called for you, so the Connections pane should automatically start up. Assign the output of pin_get() to a variable, such as con. The variable will work just like any connection variable.
The Connections Pane does not open by default when pulled via a pin. To open it use connection_view()
The con variable is now a regular database connection variable.
db_mtcars <- tbl(con, "mtcars") %>%
group_by(am) %>%
summarise(avg_mpg = mean(mpg, na.rm = TRUE))
db_mtcars
#> # Source: lazy query [?? x 2]
#> # Database: sqlite 3.29.0 [/home/edgar/connections/local.sqlite]
#> am avg_mpg
#> <dbl> <dbl>
#> 1 0 17.1
#> 2 1 24.4dplyr database queryWhen dplyr works with database data, the resulting query is not executed until the data is explicitly collected into R, or when printing the top results to the R Console. The pin records two things:
dplyr R object that contains all of the transformations. It does not save the actual results.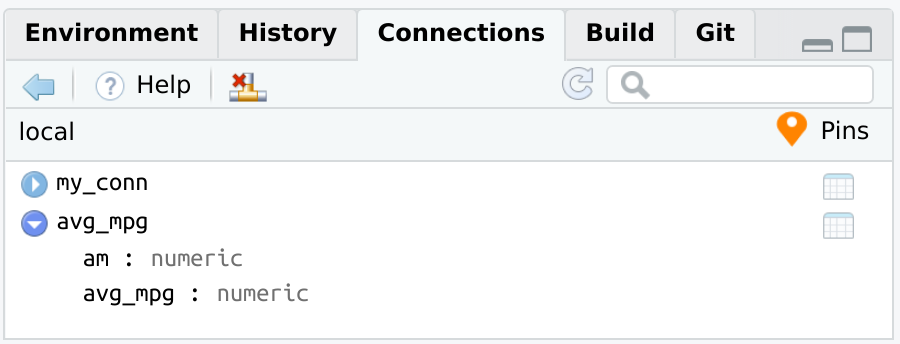
pin_get() will connect to the database, and return the dplyr object. Without assigning it to a variable, the pin will immediately print the results of the database. Those results are being processed at the time pin_get() runs.
pin_get("avg_mpg", board = "local")
#> # Source: lazy query [?? x 2]
#> # Database: sqlite 3.29.0 [/home/edgar/connections/local.sqlite]
#> am avg_mpg
#> <dbl> <dbl>
#> 1 0 17.1
#> 2 1 24.4pins exampleThe way pins integrates with databases, via the connections package, allows to open the connection from a pin, and pipe all of the subsequent code into a new pin. Afterwards, that pin can be used to collect or to continue using the dplyr object.
pin_get("my_conn", board = "local") %>%
tbl("mtcars") %>%
group_by(cyl) %>%
summarise(avg_mpg = mean(mpg, na.rm = TRUE)) %>%
pin("cyl_mpg", board = "local")
pin_get("cyl_mpg", board = "local")
#> # Source: lazy query [?? x 2]
#> # Database: sqlite 3.29.0 [/home/edgar/connections/local.sqlite]
#> cyl avg_mpg
#> <dbl> <dbl>
#> 1 4 26.7
#> 2 6 19.7
#> 3 8 15.1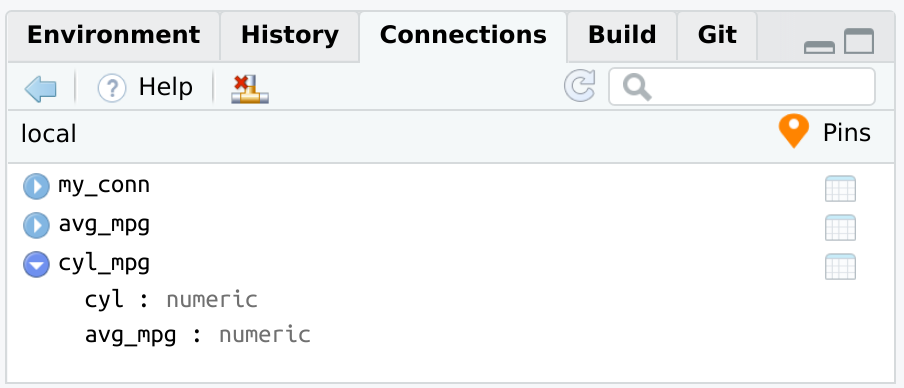
There are a couple of examples of how the Connections pane will look when opening the connection via connections.
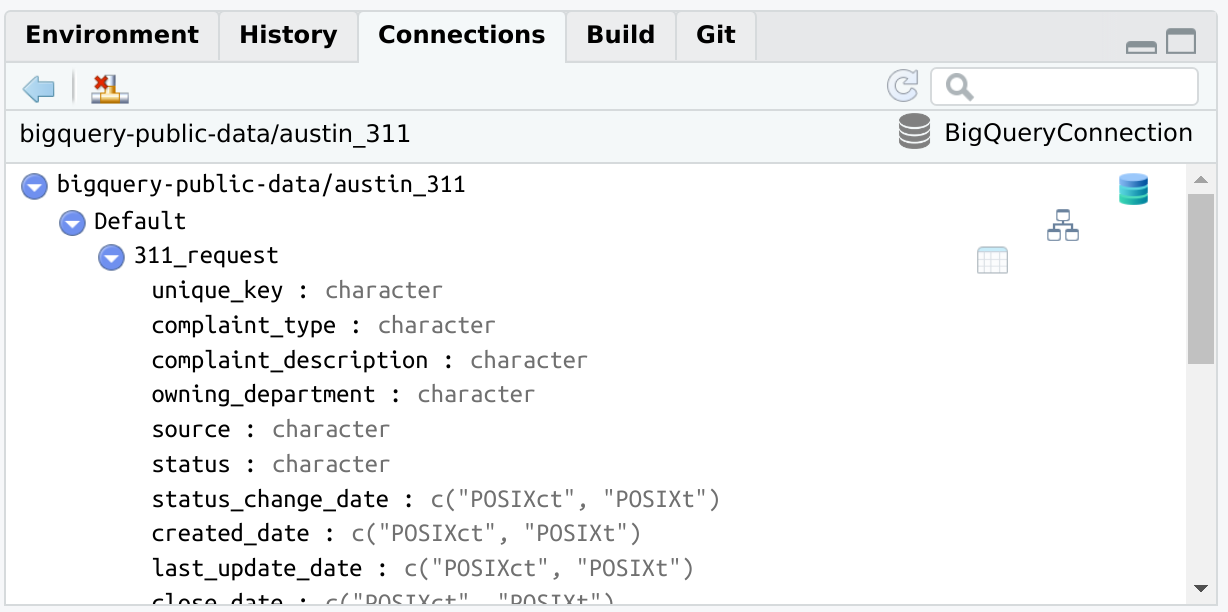
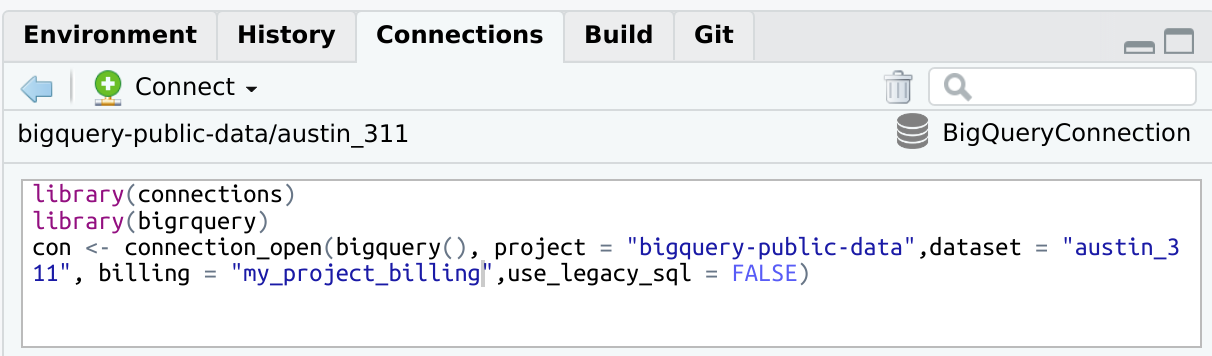
bigrquerylibrary(connections)
library(bigrquery)
con <- connection_open(
bigquery(),
project = "bigquery-public-data",
dataset = "austin_311",
billing = "my_project_billing",
use_legacy_sql = FALSE
)
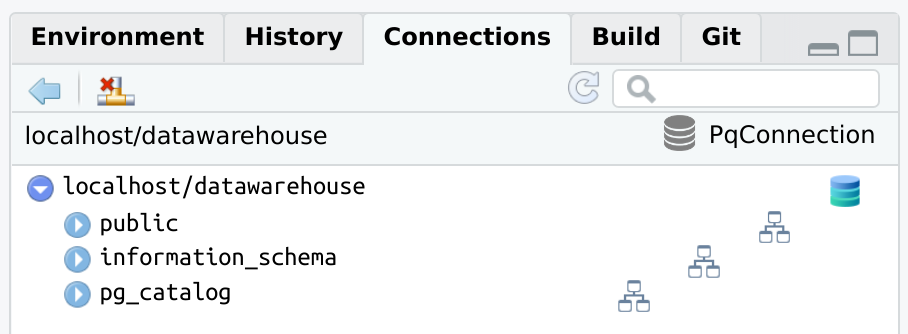
RPostgreslibrary(connections)
library(RPostgres)
con <- connection_open(Postgres(),
host = "localhost",
dbname = "datawarehouse",
user = "[user id]",
password = "[password]",
bigint = "integer",
port = "5432"
)
DBI connectionsIt is possible to integrate DBI connections not opened via connection_open(). To do that, use connection_view() and pass it the variable containing the existing database connection.
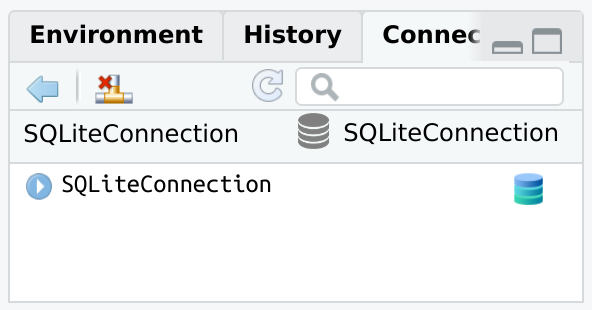
Changes to the database will not automatically load in the Connections pane. The connection_update() function will refresh the pane with the latest.