The goal of MBNMAdose is to provide a collection of useful commands that allow users to run dose-response Model-Based Network Meta-Analyses (MBNMA). This allows evidence synthesis of studies that compare multiple doses of different agents in a way that can account for the dose-response relationship.
Whilst making use of all the available evidence in a statistically robust and biologically plausible framework, this also can help connect networks at the agent level that may otherwise be disconnected at the dose/treatment level, and help improve precision of estimates. It avoids “lumping” of doses that is often done in standard Network Meta-Analysis (NMA). All models and analyses are implemented in a Bayesian framework, following an extension of the standard NMA methodology presented by (Lu and Ades 2004) and are run in JAGS (JAGS Computer Program 2017). For full details of dose-response MBNMA methodology see Mawdsley et al. (2016). Throughout this package we refer to a treatment as a specific dose or a specific agent.
On CRAN you can easily install the current release version of MBNMAdose from CRAN with:
For the development version the package can be installed directly from GitHub using the devtools R package:
# First install devtools
install.packages("devtools")
# Then install MBNMAdose directly from GitHub
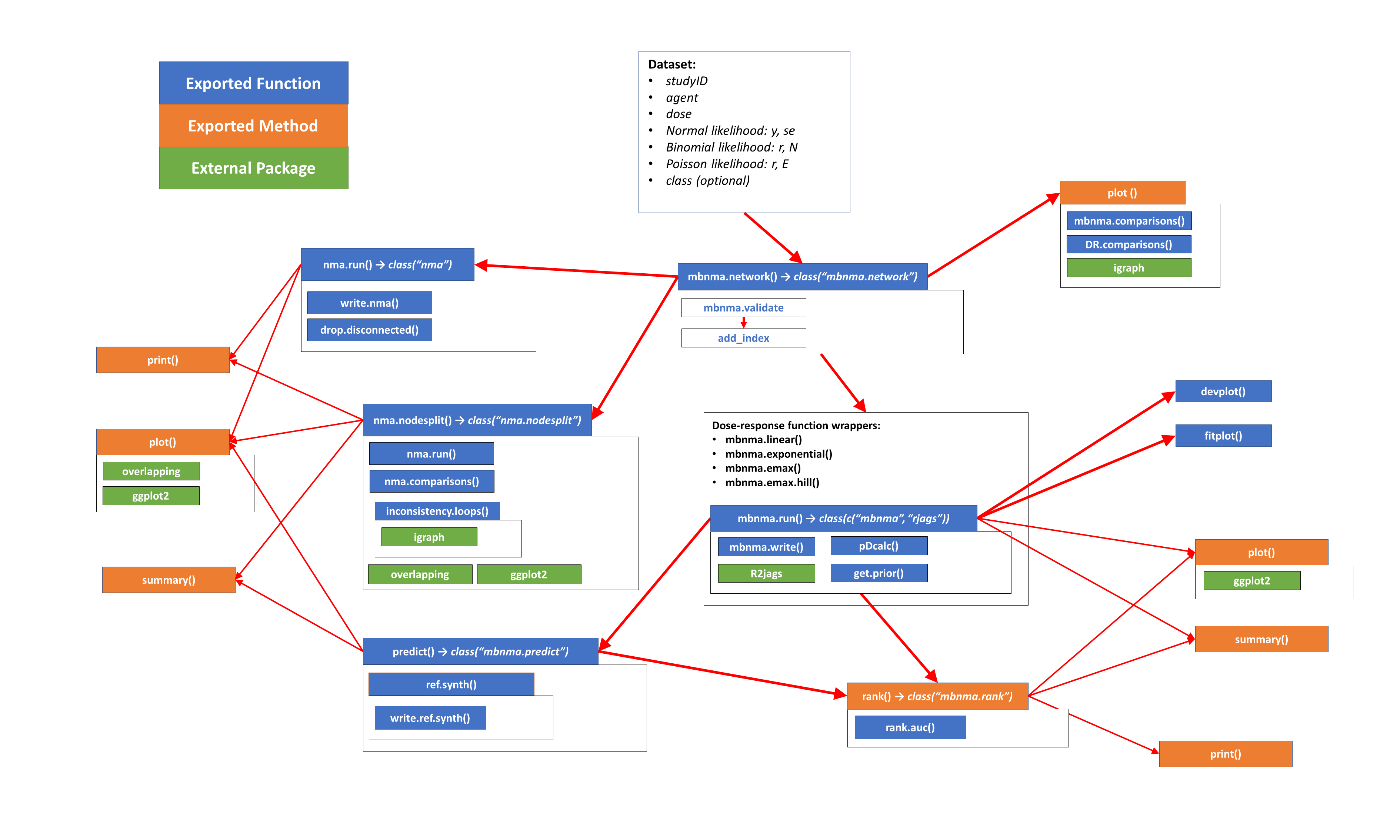
devtools::install_github("hugaped/MBNMAdose")Functions within MBNMAdose follow a clear pattern of use:
mbnma.network()mbnma.run(), or any of the available wrapper dose-response functionsnma.nodesplit() and nma.run()predict()At each of these stages there are a number of informative plots that can be generated to help understand the data and to make decisions regarding model fitting. Exported functions in the package are connected like so:

JAGS Computer Program. 2017. http://mcmc-jags.sourceforge.net.
Lu, G., and A. E. Ades. 2004. “Combination of Direct and Indirect Evidence in Mixed Treatment Comparisons.” Journal Article. Stat Med 23 (20): 3105–24. https://doi.org/10.1002/sim.1875.
Mawdsley, D., M. Bennetts, S. Dias, M. Boucher, and N. J. Welton. 2016. “Model-Based Network Meta-Analysis: A Framework for Evidence Synthesis of Clinical Trial Data.” Journal Article. CPT Pharmacometrics Syst Pharmacol 5 (8): 393–401. https://doi.org/10.1002/psp4.12091.