AlphaPart implements the method for partitioning genetic trend. The method was originally described in García-Cortés et al. (2008). The method and the package are valuable for deciphering and quantifying the sources of genetic gain. The package streamlines such analysis into a few lines of R code, while enabling advanced handling of data and results, and plotting.
You can install the released version of AlphaPart from CRAN with:
And the development version from GitHub with:
This is a basic example which shows you how to solve a common problem:
library(AlphaPart)
#>
#> Attaching package: 'AlphaPart'
#> The following object is masked from 'package:utils':
#>
#> write.csv
## Partition breeding values of a demo dataset AlphaPart.ped
## Partition the breeding values for trait 1 by "country" variable into domestic and import contributions
part <- AlphaPart(x = AlphaPart.ped, colBV = "bv1", colPath = "country")
#>
#> Size:
#> - individuals: 8
#> - traits: 1 (bv1)
#> - paths: 2 (domestic, import)
#> - unknown (missing) values:
#> bv1
#> 0
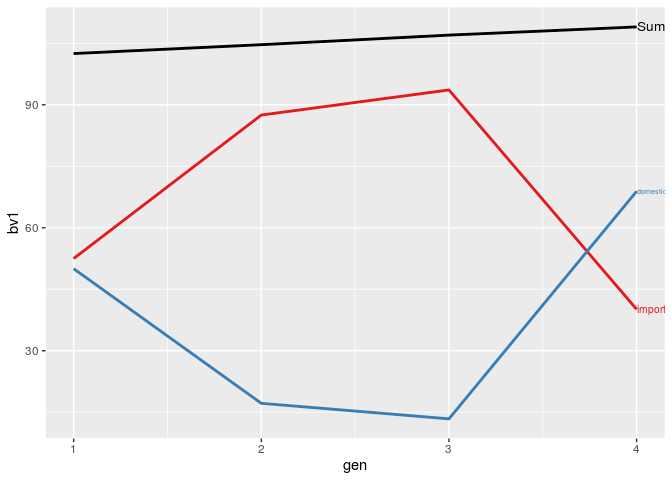
## Sumarize the partition by generation ("gen")
partSum <- summary(part, by = "gen")
## Visualize the summarized partitions